GgaEX0016692 @ galGal3
Exon Skipping
Gene
ENSGALG00000011131 | TANK
Description
NA
Coordinates
chr7:23047745-23059914:-
Coord C1 exon
chr7:23059802-23059914
Coord A exon
chr7:23052003-23052182
Coord C2 exon
chr7:23047745-23047863
Length
180 bp
Sequences
Splice sites
3' ss Seq
TTTCTAAAATTGTTTTTTAGGAA
3' ss Score
10.24
5' ss Seq
GAGGTAAGA
5' ss Score
10.06
Exon sequences
Seq C1 exon
GTAATATGCATCCCTACATTCTGAGGCATGAAGATATTGAAACCTCCAACTTATCTTTTAGCCAGCTCTCTGAAAAACTGAATGTAGCAAAGCGAAGAGAAAGACTCCTAAAG
Seq A exon
GAACAGTTAGAAAATGAGAGCATGAAGTTGAAGCAACTTGAGGACAGAAACAGTGAAAAGGAAAGGAGGCTTATGTCTATTGTTTCCAACCAAGAAGATAAAATACGAACTCTGAAAAACAAATTGAAAGAATTAAGTGACGCCCAAAAGGGTACACAGATGCCAACATACAAAATAGAG
Seq C2 exon
GGAGAATCTGGAGACTATTTTTCAGGAGATGAAAGAAGAGTGTCATCGAATATGTATGCTAGCCAGAGAACAAACAGACCAACTGAGTAAATTTAAAATAAAGCCAGAACCTGAAACTG
VastDB Features
Vast-tools module Information
Secondary ID
ENSGALG00000011131_MULTIEX1-1/3=C1-3
Average complexity
C2
Mappability confidence:
NA
Protein Impact
Alternative protein isoforms (No Ref)
Show structural model
Features
Disorder rate (Iupred):
C1=0.151 A=0.700 C2=0.341
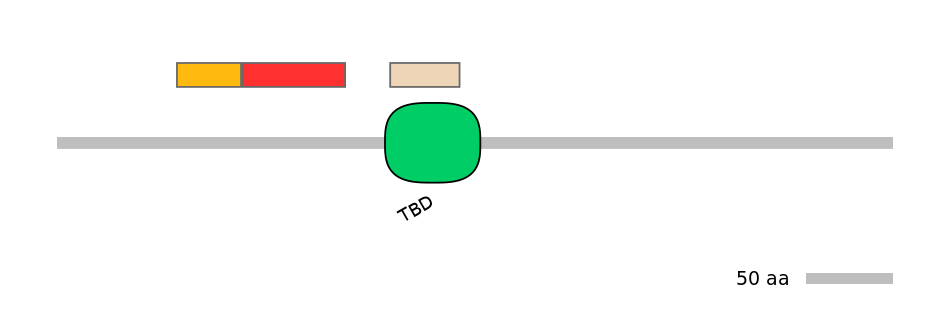
Domain overlap (PFAM):
C1:
PF083176=Spc7=FE(32.5=100),PF0192313=Cob_adeno_trans=FE(32.5=100),PF0246513=FliD_N=FE(37.8=100),PF103924=COG5=FE(34.6=100)
A:
NO
C2:
PF128452=TBD=FE(71.4=100)
Other Inclusion Isoforms:
NA
Associated events
Other assemblies
Conservation
Human
(hg38)
No conservation detected
Human
(hg19)
No conservation detected
Mouse
(mm10)
No conservation detected
Mouse
(mm9)
No conservation detected
Rat
(rn6)
No conservation detected
Cow
(bosTau6)
No conservation detected
Zebrafish
(danRer10)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
TGCATCCCTACATTCTGAGGC
R:
CAGTTTCAGGTTCTGGCTTTATT
Band lengths:
226-406
Functional annotations
There are 0 annotated functions for this event