GgaEX0018894 @ galGal3
Exon Skipping
Gene
ENSGALG00000012495 | Q9DEH4_CHICK
Description
NA
Coordinates
chr7:36827767-36831912:-
Coord C1 exon
chr7:36831814-36831912
Coord A exon
chr7:36830453-36830551
Coord C2 exon
chr7:36827767-36827877
Length
99 bp
Sequences
Splice sites
3' ss Seq
TCTCTTGCTACGCGTTATAGGCG
3' ss Score
5.25
5' ss Seq
CATGTAAGA
5' ss Score
5.58
Exon sequences
Seq C1 exon
ATTGAATATAAAAAGCGTGGAGAAGCTGGCCTGGGTGTAACCATGCTGGGACGTCCAGATATTGAGCTGGCAAAGGAAGTGTCCAAGCTAACCAGTCAG
Seq A exon
GCGGAATACCTGAAGCGGCATATGAGAGGGCACGGAGCTGCATCTTATGATACACCATGGATGAGAACGTTTAAGAAAGCTAATATGCTAAGTAGTCAT
Seq C2 exon
GTGAAGTACAAGGAGAACTTTTCCAAAGAGAAGGGCAAAAAGCCAAAGTATGATTTAAAGGAGGCTAAAATCTACAAGACCATGAAAGATGCTCACAACATAGCTAGTGAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSGALG00000012495-'127-127,'127-126,128-127
Average complexity
S
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (No Ref)
No structure available
Features
Disorder rate (Iupred):
C1=0.000 A=0.182 C2=0.135
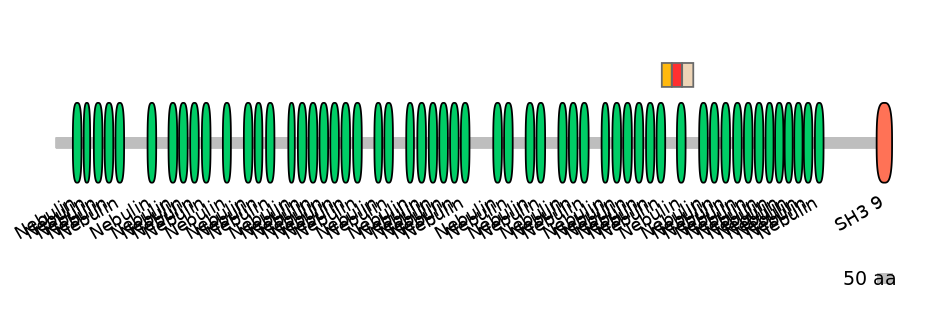
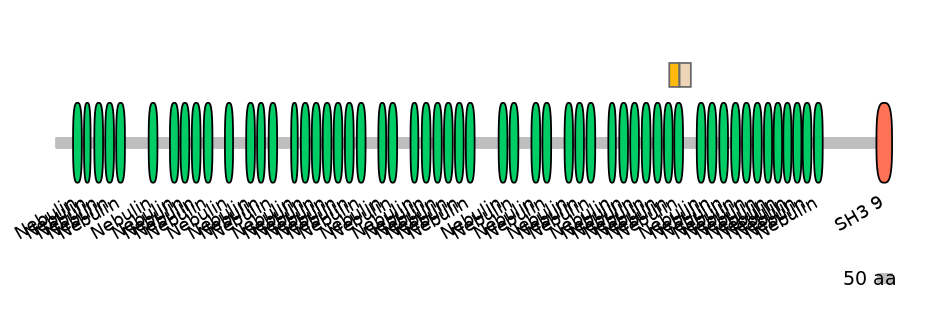
Domain overlap (PFAM):
C1:
PF0088013=Nebulin=PD(37.9=33.3),PF0088013=Nebulin=PU(55.2=48.5)
A:
PF0088013=Nebulin=PU(55.2=48.5)
C2:
PF0088013=Nebulin=PD(37.9=29.7)
Other Inclusion Isoforms:
NA
Associated events
Other assemblies
Conservation
Human
(hg38)
No conservation detected
Mouse
(mm10)
No conservation detected
Mouse
(mm9)
No conservation detected
Rat
(rn6)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
GTGTAACCATGCTGGGACGTC
R:
CTCACTAGCTATGTTGTGAGCA
Band lengths:
176-275
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]