MmuEX0016483 @ mm10
Exon Skipping
Gene
ENSMUSG00000013787 | Ehmt2
Description
euchromatic histone lysine N-methyltransferase 2 [Source:MGI Symbol;Acc:MGI:2148922]
Coordinates
chr17:34905342-34906058:+
Coord C1 exon
chr17:34905342-34905456
Coord A exon
chr17:34905610-34905711
Coord C2 exon
chr17:34905910-34906058
Length
102 bp
Sequences
Splice sites
3' ss Seq
TCTCTCTGTCTCTGTGTCAGGAG
3' ss Score
8.74
5' ss Seq
CTGGTAATT
5' ss Score
5.99
Exon sequences
Seq C1 exon
AGCGGTTCTAGTGGCCGGCGCAAGGCCAAGAAGAAATGGCGGAAAGACAGCCCGTGGGTGAAGCCATCTAGAAAACGGCGGAAACGAGAGCCTCCGAGGGCCAAGGAGCCAAGAG
Seq A exon
GAGTGAATGGTGTGGGTTCCTCAGGGCCCAGTGAGTACATGGAGGTTCCTCTGGGGTCCCTGGAGCTGCCCAGCGAGGGGACCCTCTCCCCCAACCACGCTG
Seq C2 exon
GGGTCTCCAATGACACGTCTTCACTGGAGACAGAACGCGGGTTTGAGGAGCTGCCCCTCTGCAGCTGCCGCATGGAGGCTCCCAAGATTGACCGCATCAGCGAGAGAGCAGGGCACAAGTGCATGGCCACAGAGAGTGTGGATGGAGAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSMUSG00000013787_CASSETTE2
Average complexity
S
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (Ref)
No structure available
Features
Disorder rate (Iupred):
C1=0.731 A=1.000 C2=0.233
Domain overlap (PFAM):
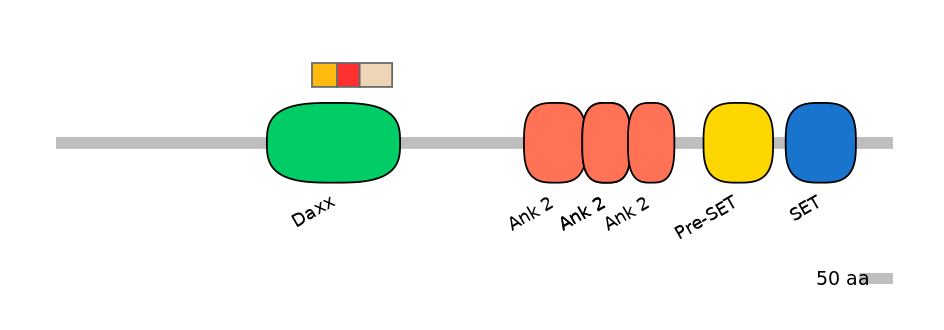
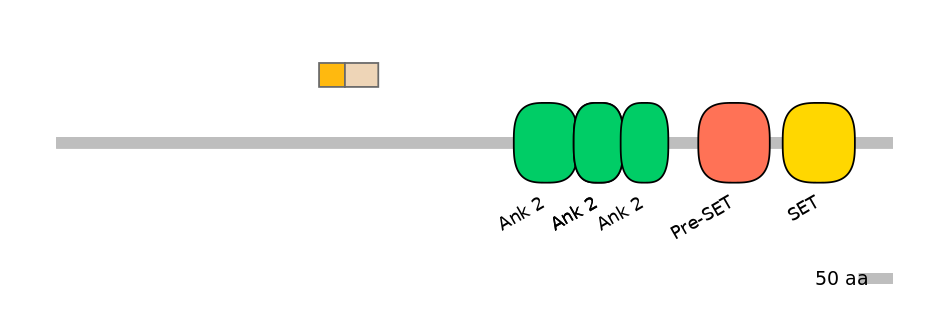
C1:
PF0334410=Daxx=FE(18.8=100)
A:
PF0334410=Daxx=FE(16.8=100)
C2:
PF0334410=Daxx=FE(24.3=100)
Associated events
Other assemblies
Conservation
Chicken
(galGal3)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
GCGCAAGGCCAAGAAGAAATG
R:
CTCTCCATCCACACTCTCTGT
Band lengths:
247-349
Functional annotations
There are 2 annotated functions for this event
PMID: 26997278
This event
Although E10 inclusion greatly stimulates overall H3K9me2 levels, it does not affect G9a catalytic activity. Instead, E10 increases G9a nuclear localization. The authors show that the G9a E10(+) isoform is necessary for neuron differentiation and regulates the alternative splicing pattern of its own pre-mRNA, enhancing E10 inclusion.
PMID: 11707778
The sub-cellular distribution of the NG36/G9a-T7 and G9a-T7 proteins was found to be quite distinct. Whereas the G9a-T7 protein was observed in both the cytoplasm and the nucleus, the NG36/G9a-T7 protein was extensively concentrated within the nucleus. Also, the G9a-T7 protein frequently appeared marginalized at the nuclear periphery, while the NG36/G9a-T7 protein was generally found throughout the nucleoplasm.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Pre-implantation embryo development
- Ribosome-engaged transcriptomes of neuronal types
- Neural differentiation time course
- Muscular differentiation time course
- Spermatogenesis cell types
- Reprogramming of fibroblasts to iPSCs
- Hematopoietic precursors and cell types