RnoEX0031276 @ rn6
Exon Skipping
Gene
ENSRNOG00000030630 | Ehmt2
Description
euchromatic histone lysine methyltransferase 2 [Source:RGD Symbol;Acc:1302972]
Coordinates
chr20:4583857-4584570:+
Coord C1 exon
chr20:4583857-4583971
Coord A exon
chr20:4584122-4584223
Coord C2 exon
chr20:4584422-4584570
Length
102 bp
Sequences
Splice sites
3' ss Seq
TCTCTCTGTCTCTGTGTCAGGAG
3' ss Score
8.74
5' ss Seq
CTGGTAATT
5' ss Score
5.99
Exon sequences
Seq C1 exon
AGTGGCTCTAGCGGCCGACGCAAGGCCAAGAAGAAATGGCGGAAAGACAGCCCATGGGTGAAGCCATCCAGGAAACGGCGGAAGCGAGAGCCTCCGAGGGCCAAGGAGCCCAGAG
Seq A exon
GAGTGAATGGTGTGGGTTCCTCAGGGCCCAGTGAGTACATGGAGGTTCCTCTGGGGTCCCTGGAGCTGCCCAGCGAGGGGACCCTCTCCCCCAACCACGCTG
Seq C2 exon
GGGTCTCCAATGACACGTCTTCGCTGGAGACGGAGCGTGGGTTTGAGGAGCTGCCCCTCTGCAGCTGCCGCATGGAGGCGCCCAAGATTGACCGCATCAGCGAGAGAGCAGGACACAAGTGTATGGCCACCGAAAGTGTGGACGGAGAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSRNOG00000030630_CASSETTE3
Average complexity
S*
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (Ref)
No structure available
Features
Disorder rate (Iupred):
C1=0.733 A=1.000 C2=0.232
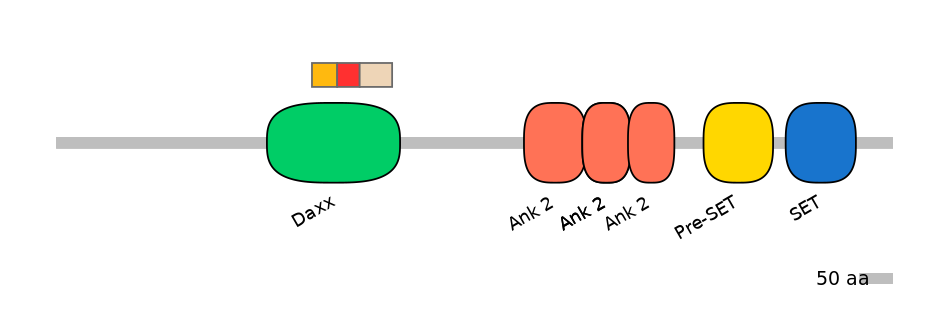
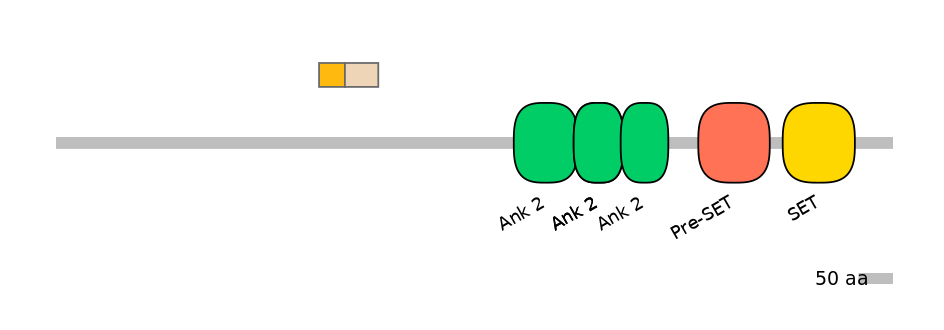
Domain overlap (PFAM):
C1:
PF0334410=Daxx=FE(18.8=100)
A:
PF0334410=Daxx=FE(16.8=100)
C2:
PF0334410=Daxx=FE(24.3=100)
Associated events
Conservation
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
CGCAAGGCCAAGAAGAAATGG
R:
CTCTCCGTCCACACTTTCGGT
Band lengths:
246-348
Functional annotations
There are 2 annotated functions for this event
PMID: 11707778
The sub-cellular distribution of the NG36/G9a-T7 and G9a-T7 proteins was found to be quite distinct. Whereas the G9a-T7 protein was observed in both the cytoplasm and the nucleus, the NG36/G9a-T7 protein was extensively concentrated within the nucleus. Also, the G9a-T7 protein frequently appeared marginalized at the nuclear periphery, while the NG36/G9a-T7 protein was generally found throughout the nucleoplasm.
PMID: 26997278
Although E10 inclusion greatly stimulates overall H3K9me2 levels, it does not affect G9a catalytic activity. Instead, E10 increases G9a nuclear localization. The authors show that the G9a E10(+) isoform is necessary for neuron differentiation and regulates the alternative splicing pattern of its own pre-mRNA, enhancing E10 inclusion.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]