MmuEX0017049 @ mm9
Exon Skipping
Gene
ENSMUSG00000024240 | Epc1
Description
enhancer of polycomb homolog 1 (Drosophila) [Source:MGI Symbol;Acc:MGI:1278322]
Coordinates
chr18:6441120-6449255:-
Coord C1 exon
chr18:6448903-6449255
Coord A exon
chr18:6441800-6441910
Coord C2 exon
chr18:6441120-6441241
Length
111 bp
Sequences
Splice sites
3' ss Seq
TTTTTGTTTGTCTTTTTTAGAAC
3' ss Score
12.32
5' ss Seq
CAGGTGGGT
5' ss Score
8.56
Exon sequences
Seq C1 exon
GGTCGTGCTGGACAGAGCGCATTCAGACTATGACAGTATGTTTCACCACCTGGATTTGGACATGCTTTCCTCACCACAACCTTCTCCAGTCAATCAGTTTGCCAATACCTCAGAACCCAATACCTCGGACAGATCTTCCTCTAAAGACCTCAGTCAGATACTAGTCGATATCAAATCATGTAGATGGCGGCACTTTAGGCCTCGGACACCATCCCTGCCTGACAGTGACAGTGGTGAACTCTCCAGTAGAAAGTTACACAGGAGTATCAGTCGAGCAGGAGCAGCACAGCCTGGGGCCCACACGTGCAGCACCTCCACCCAGAACAGAAGTAGCAGTGGCTCAGCACACTGTG
Seq A exon
AACTGAGCTCTCCATAGACACACATAGTAGCATAACACAGTGTCTGTCCTCAGGCTTCACTCTGTACAGTAATCAGCATAACTCCGTTCTTATTGACTGGATTTTGTCCAG
Seq C2 exon
CATTCACAGCCGAACAGTACCAGCAGCACCAGCAGCAGCTGGCACTCATGCAGCAGCAGCAGCTTGCGCAGACTCAGCAGCAGCAGCAAGCAAATAGTAGTTCCTCTGCCGCCGCGCAACAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSMUSG00000024240-'13-15,'13-13,14-15
Average complexity
S
Mappability confidence:
100%=100=100%
Protein Impact
ORF disruption upon sequence inclusion
No structure available
Features
Disorder rate (Iupred):
C1=0.697 A=1.000 C2=0.794
Domain overlap (PFAM):
C1:
PF067527=E_Pc_C=PU(0.1=0.0)
A:
NO
C2:
PF067527=E_Pc_C=PU(17.2=97.6)
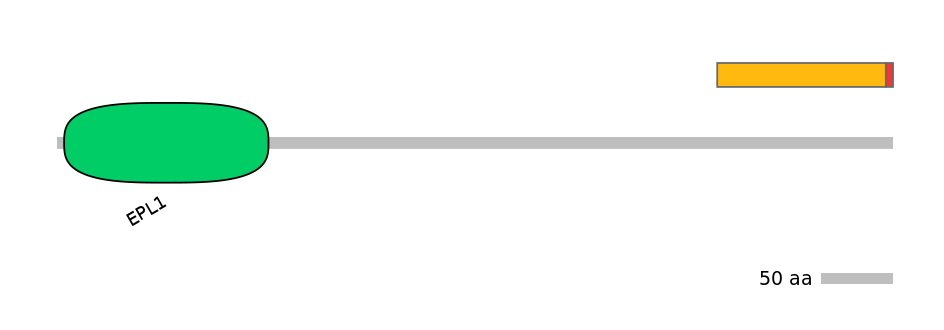
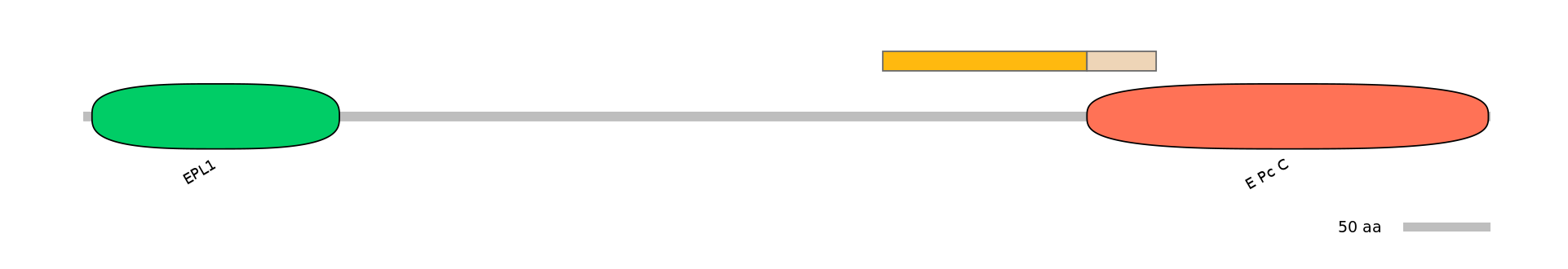
Other Inclusion Isoforms:
NA
Associated events
Other assemblies
Conservation
Chicken
(galGal4)
No conservation detected
Chicken
(galGal3)
No conservation detected
Zebrafish
(danRer10)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
CTTTAGGCCTCGGACACCATC
R:
CTATTTGCTTGCTGCTGCTGC
Band lengths:
258-369
Functional annotations
There are 2 annotated functions for this event
PMID: 31911676
CRISPR targeting of the poised exons showed a modest fitness advantage in cell culture. Tumors derived from engraftment of polyclonal EPC1 poison exon targeted PC9-Cas9 cells were significantly larger and exhibited increased Ki-67 staining relative to control tumors.
PMID: 31911676
[CRISPR screen]. Conserved poison exon with pro-oncogenic impact when depleted: HeLa No Change at 14 days (FC=1.187, FDR=0.002), PC9 No Change at 14 days (FC=1.144, FDR=0.102), Late Xenograft Enriched (FC=2.344, FDR=0.000).
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Pre-implantation embryo development
- Neural differentiation time course
- Muscular differentiation time course
- Spermatogenesis cell types
- Reprogramming of fibroblasts to iPSCs
- Hematopoietic precursors and cell types
Other AS DBs: