MmuEX0039821 @ mm9
Exon Skipping
Gene
ENSMUSG00000036202 | Rif1
Description
Rap1 interacting factor 1 homolog (yeast) [Source:MGI Symbol;Acc:MGI:1098622]
Coordinates
chr2:51929767-51931697:+
Coord C1 exon
chr2:51929767-51929845
Coord A exon
chr2:51930750-51930838
Coord C2 exon
chr2:51931601-51931697
Length
89 bp
Sequences
Splice sites
3' ss Seq
GTTACTATTTTTTTTGACAGGGC
3' ss Score
7.13
5' ss Seq
CAGGCACGT
5' ss Score
2.9
Exon sequences
Seq C1 exon
TCGTATGACTGGAGAAGAAGGAAAAGAAGTCATTGCAGAAATCGAGAAAAACCTTTCTCGGCTGTACACGGTTTTAAAG
Seq A exon
GGCCTCTAGCCCAGGCTGGCCTTGGACTTGTATATAGCTGAGGATGACCCTGACCTGATCCTTCTGCCTCCCAAGTGCTGGGATCACAG
Seq C2 exon
GCTCACATTTCCAGTCAGAACTCAGAACTGAGCAGTGCTGCTTTGCAAGCCTTGGGATTTTGTTTATATAATCCCAGAATTACCTCAGGATTGTCAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSMUSG00000036202_MULTIEX1-2/3=1-3
Average complexity
C1
Mappability confidence:
100%=100=100%
Protein Impact
ORF disruption upon sequence inclusion
No structure available
Features
Disorder rate (Iupred):
C1=0.085 A=0.000 C2=0.000
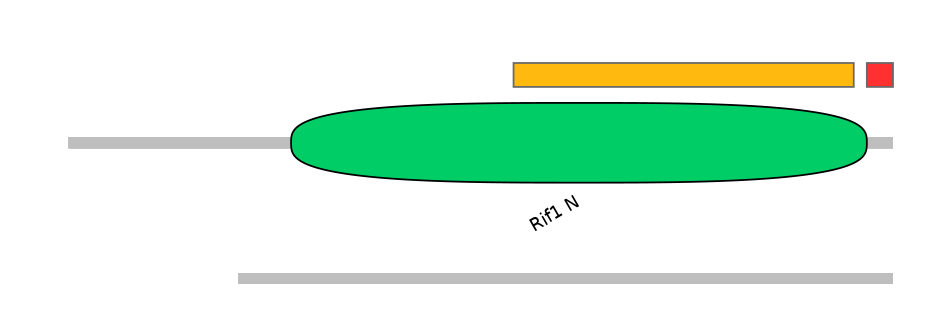
Domain overlap (PFAM):
C1:
PF122313=Rif1_N=FE(57.8=100),PF074086=DUF1507=FE(76.5=100)
A:
PF122313=Rif1_N=PD(0.1=0.0),PF074086=DUF1507=PD(0.1=0.0)
C2:
PF122313=Rif1_N=FE(8.8=100)
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
Associated events
Other assemblies
Conservation
Human
(hg38)
No conservation detected
Human
(hg19)
No conservation detected
Rat
(rn6)
No conservation detected
Cow
(bosTau6)
No conservation detected
Chicken
(galGal4)
No conservation detected
Chicken
(galGal3)
No conservation detected
Zebrafish
(danRer10)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
TCGTATGACTGGAGAAGAAGGA
R:
TGACAATCCTGAGGTAATTCTGGG
Band lengths:
175-264
Functional annotations
There are 1 annotated functions for this event
PMID: 34736895
Ectopic expression of Rif1 lacking N-terminal domain results in upregulation of 2C transcripts. This appears to be caused by dominant negative inhibition of endogenous Rif1 protein localization at the nuclear periphery through formation of hetero-oligomers between the N-terminally truncated and endogenous forms. In murine 2-cell embryos, most of Rif1-derived polypeptides are expressed as truncated forms in soluble nuclear or cytosolic fraction, and are likely non-functional. Toward the morula stage, the full-length form of Rif1 gradually increased. Note: this exon is one possible isoform that is truncated at the N-term; however, this paper does not show any specific event.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Pre-implantation embryo development
- Neural differentiation time course
- Muscular differentiation time course
- Spermatogenesis cell types
- Reprogramming of fibroblasts to iPSCs
- Hematopoietic precursors and cell types
Other AS DBs: