DreEX0003172 @ danRer10
Exon Skipping
Gene
ENSDARG00000009026 | CR388002.1
Description
Uncharacterized protein [Source:UniProtKB/TrEMBL;Acc:E7F4G7]
Coordinates
chr1:13433709-13438999:+
Coord C1 exon
chr1:13433709-13433933
Coord A exon
chr1:13438339-13438437
Coord C2 exon
chr1:13438845-13438999
Length
99 bp
Sequences
Splice sites
3' ss Seq
AATGGCTGCGATCGGAACAGTAA
3' ss Score
2.18
5' ss Seq
CGGGTAGGA
5' ss Score
7.22
Exon sequences
Seq C1 exon
CTTCCTGGTCAGTTTTATGGTGGATGCTCGTGGTGGAGCCATGCGTGGCTGTCGTCACAATGGACTACGCATTATTGTTCCACCTAGAAAGTGTTCAGCTCCTACACGAGTCACGTGTCGCCTGGTGAAGAGGCACAGACTGGCCTCTATGCCCCCAATGGTGGAAGGAGAGGGGCTTGCTGGCAAAATCATTGAAGTTGGTCCCACCGGAGCACAGTTTCTTGG
Seq A exon
TAAACTGCACCTCCCAACAGCCCCTCCTCCTCTAAATGAGGGTGAAAGTCTTGTCAGCAGAATTCTGCAGTTGGGTCCACCCGGCACCAAATTCCTCGG
Seq C2 exon
GCCTGTAATAGTGGAGATCCCGCACTTCGCTGCATTGCGTGGGACAGAGAGAGAGTTAGTGATTCTGCGGAGTGAGACGGGAGAGAGCTGGAGGGAACATCACTGTGAGCATACAGAAGAAGAGCTCAATCAGATACTCAATGGCATGGATGAGG
VastDB Features
Vast-tools module Information
Secondary ID
ENSDARG00000009026_CASSETTE3
Average complexity
S
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (Ref)
No structure available
Features
Disorder rate (Iupred):
C1=0.079 A=0.353 C2=0.434
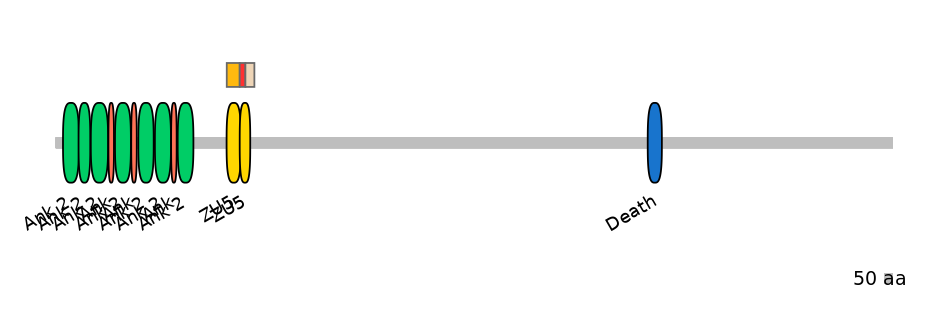
Domain overlap (PFAM):
C1:
PF0079115=ZU5=FE(89.3=100),PF0079115=ZU5=PU(0.1=0.0)
A:
PF0079115=ZU5=PD(8.3=20.6),PF0079115=ZU5=PU(53.2=97.1)
C2:
PF0079115=ZU5=PD(45.2=52.8)
Main Skipping Isoform:
NA
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Conservation
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
CTGGTGAAGAGGCACAGACTG
R:
CGTCTCACTCCGCAGAATCAC
Band lengths:
183-282
Functional annotations
There are 1 annotated functions for this event
PMID: 22749401
Inclusion enhances interaction with EPB4L2, DCTN4, SCN1B, SLC8A1, DNAJB1, CDH1, ATP1A1, SPNA2. By LUMIER.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]