DreEX0044955 @ danRer10
Exon Skipping
Gene
ENSDARG00000091777 | mapkap1
Description
mitogen-activated protein kinase associated protein 1 [Source:ZFIN;Acc:ZDB-GENE-041111-178]
Coordinates
chr5:4972798-4993970:+
Coord C1 exon
chr5:4972798-4972907
Coord A exon
chr5:4990094-4990201
Coord C2 exon
chr5:4993830-4993970
Length
108 bp
Sequences
Splice sites
3' ss Seq
TTCAGTGTTTTGGTTTTTAGGCC
3' ss Score
8.58
5' ss Seq
ACAGTGAGT
5' ss Score
8.34
Exon sequences
Seq C1 exon
AAACGCAGCTCATGGTTTCTCCTTGATCCCGGTGGACAGTCTGAAAGTGACCATGAAGGATATCCTGCAGAAAGCCCTCAAGAAGAGGAAGGGCTCCCAGAAAGGATCAG
Seq A exon
GCCCCTTGTACCGACTGGAGAAGCAAATGGAGCCGAATGTGGCGGTGGATCTGGACAGTTCGCTGGAGAACCAGGGAACGCTGGAGTTCTGCTTGGTCCGAGAAAACA
Seq C2 exon
GCTCTCGAGGCGAGGAGAGCTCAGAGGAGGATCCACCTATAGACATCACTACTGTGCAGGACATGTTGAGCAGTCACCACTACAAGTCCTTCAAGATCAGCATGATCCACAAGCTCCGCTTCACCACAGACGTGCAGCTAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSDARG00000091777_CASSETTE1
Average complexity
S
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (No Ref)
No structure available
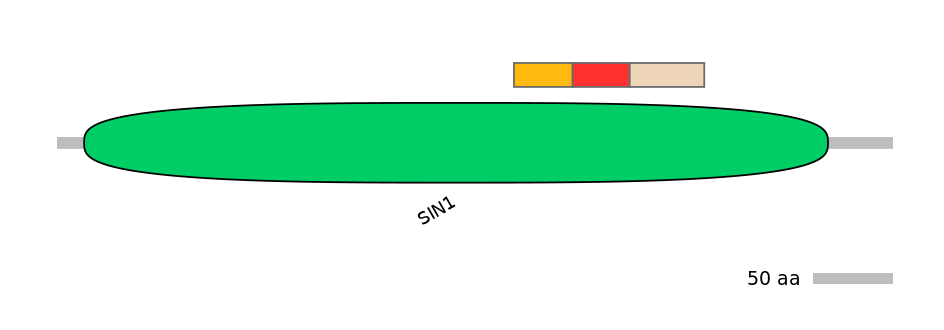
Features
Disorder rate (Iupred):
C1=0.053 A=0.324 C2=0.333
Domain overlap (PFAM):
C1:
PF054227=SIN1=FE(7.9=100)
A:
PF054227=SIN1=FE(7.7=100)
C2:
PF054227=SIN1=FE(10.0=100)
Main Skipping Isoform:
NA
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Conservation
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
CGCAGCTCATGGTTTCTCCTT
R:
TAGCTGCACGTCTGTGGTGAA
Band lengths:
247-355
Functional annotations
There are 1 annotated functions for this event
PMID: 16919458
[No functional differences found]. Immunoprecipitation demonstrates that mSin1.1 (all included), mSin1.2 (skipping of HsaEX0037808), and mSin1.5 (AI and APA) integrate into mTORC2 complexes. Insulin treatment increases the kinase activity of mSin1.1 and mSin1.2 but not mSin1.5 complexes.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]