DreEX0051221 @ danRer10
Exon Skipping
Gene
ENSDARG00000074697 | npdc1a
Description
neural proliferation, differentiation and control, 1a [Source:ZFIN;Acc:ZDB-GENE-031204-31]
Coordinates
chr21:13618551-13654806:-
Coord C1 exon
chr21:13654431-13654806
Coord A exon
chr21:13629375-13629527
Coord C2 exon
chr21:13618551-13618643
Length
153 bp
Sequences
Splice sites
3' ss Seq
TTTCTTCTTTTGTTTTTCAGCAA
3' ss Score
10.8
5' ss Seq
GTGGTATGA
5' ss Score
5.27
Exon sequences
Seq C1 exon
TTCCGCTGCCGCAGGGATCTCCGCTAATCTGTCCTGTCAATCAACGGAGGTGCCGCTCCTCCTCCTTTTTCCTCCTCCTAAATCTCTCTCCTCTATCTTTAGCTCTGTACCACGGCGGCTTAACGCTCTTTCAGAGTTGACGCTCCCATCTTCATTCGGGCCGGTTTGTTCTCTGTCAGTCGCGTACGTCCAACCGACCTACATCCAGATTTGGGAATGACACCCCGCGGCGTTTATCTGCTGTCCTGGACTTGAAAAGCTCCGTGAAGGATGTTGTTTTCTCCGACCCGGAGCACACGGCGAGCCCGGGCCTCCCTTCTGCTGCCCGGGCTGCTGATCTGCGTTATCCTCGCCGCCGTCGGTGCGAGTGTGCCGG
Seq A exon
CAAAGTGCCCTGATCACATAGAGTGTGCCAGAAGGGGGCGTCACTTCTGCAAGCCTGGCTCCTCCAACTGCGGCCCCTGCCTCATCCCACTGGAGGAGGATCAAATGGGACATTGTGTGGTTCGCAGTGTACGATATGCAGCTCAGTCCCGTG
Seq C2 exon
ATCCAGTGGCCCCACTTTCTGGCATAGATGAAGAAATCGACTTTCTGTCCTCCGTCATCACCAAGCAGCAGTTGTCGGAAAGCAAGCACTCAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSDARG00000074697-'1-2,'1-0,5-2
Average complexity
S
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (Ref)
No structure available
Features
Disorder rate (Iupred):
C1=0.000 A=0.000 C2=0.219
Domain overlap (PFAM):
C1:
PF068096=NPDC1=PU(10.9=91.7)
A:
PF068096=NPDC1=FE(16.8=100)
C2:
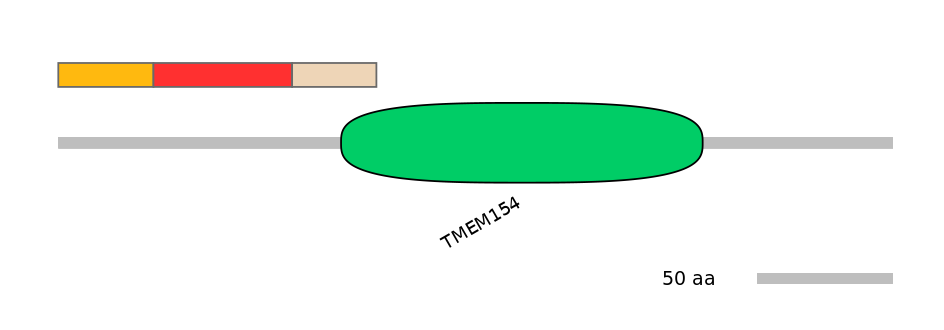
PF068096=NPDC1=FE(10.2=100),PF151021=TMEM154=PU(9.7=40.6)
Main Skipping Isoform:
NA
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Conservation
Cow
(bosTau6)
No conservation detected
Chicken
(galGal3)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
CGGCGTTTATCTGCTGTCCTG
R:
CTGAGTGCTTGCTTTCCGACA
Band lengths:
242-395
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]