DreEX0051325 @ danRer10
Exon Skipping
Gene
ENSDARG00000018721 | npnt
Description
nephronectin [Source:ZFIN;Acc:ZDB-GENE-090312-201]
Coordinates
chr1:49467822-49488185:+
Coord C1 exon
chr1:49467822-49467922
Coord A exon
chr1:49482331-49482381
Coord C2 exon
chr1:49488093-49488185
Length
51 bp
Sequences
Splice sites
3' ss Seq
TCTGGCCGTTTGTTTTGCAGCCC
3' ss Score
10.89
5' ss Seq
AAGGTTAGC
5' ss Score
5.61
Exon sequences
Seq C1 exon
GTGGTCCCGACAGATGTCCTCATCCAATGGTCTGTGTCGGTACGGCGGCCGCATTGACTGCTGCTGGGGCTGGACGAGAGTCTCCTGGGGCCAGTGCCAGC
Seq A exon
CCCTCTACGTCTTAACCCGCAGAGTAATCGGGATAAGGTGTCAGCCCCAAG
Seq C2 exon
CGCTATGTCAGCACGGATGCAAGCACGGAGAATGCGTGGGACCCAACAAATGCAAGTGCCATCCAGGTTATACAGGAAAAACATGCAACCAAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSDARG00000018721_CASSETTE1
Average complexity
S
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (Ref)
No structure available
Features
Disorder rate (Iupred):
C1=0.000 A=0.000 C2=0.000
Domain overlap (PFAM):
C1:
PF094025=MSC=FE(43.6=100)
A:
NO
C2:
PF126612=hEGF=WD(100=40.6),PF0764510=EGF_CA=PU(0.1=0.0)
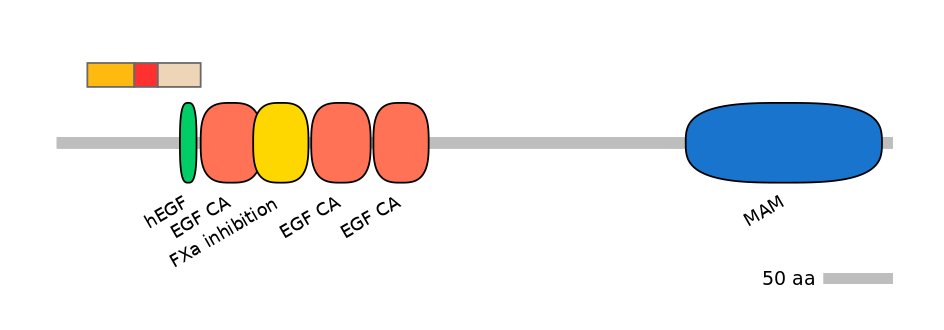
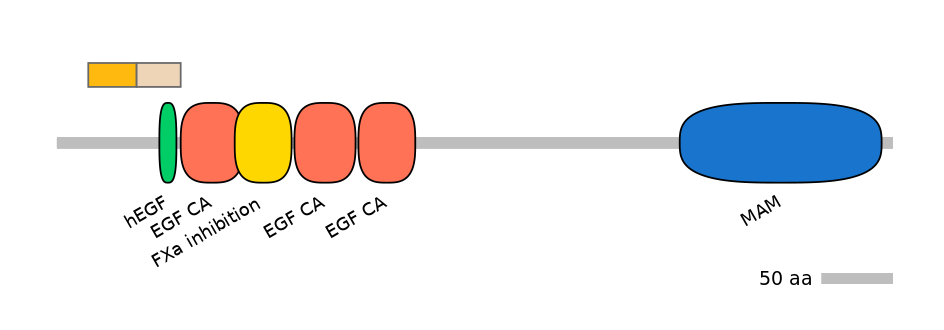
Associated events
Conservation
Cow
(bosTau6)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
CATCCAATGGTCTGTGTCGGT
R:
ATTTGTTGGGTCCCACGCATT
Band lengths:
132-183
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]