DreEX0080938 @ danRer10
Exon Skipping
Gene
ENSDARG00000002168 | tra2b
Description
transformer 2 beta homolog (Drosophila) [Source:ZFIN;Acc:ZDB-GENE-040426-1094]
Coordinates
chr9:12613331-12623556:-
Coord C1 exon
chr9:12623383-12623556
Coord A exon
chr9:12618736-12618881
Coord C2 exon
chr9:12613331-12613519
Length
146 bp
Sequences
Splice sites
3' ss Seq
CTGGGATGTGGTTGGTTTAGGAG
3' ss Score
3.58
5' ss Seq
CAGGTAAAG
5' ss Score
9.65
Exon sequences
Seq C1 exon
TTCTAAATAAAGCTTCTACGGCGAGAAAGGCGTTCATTTACAGACGTCTGGTTTGTGTGTGCGTTTGAGCACTACTTGTGTGTTGTCCTAATCATTTCTCTTGCATTGTAATTATTGCAGCATATTACAAACTAAACAAAAATGAGTGACGCCGAGAAGGAATTCGTGGAGCGG
Seq A exon
GAGTCTCGTTCAGCCTCTAGGAGCGCAAGTCCTAGAGGATCTGCCAAGTCCGGCAGTCGCTCAGCTGAACGCTCGCCTGCTCATTCCAAAGAGAGGTCCCATCATTCCCGCTCAAAATCCCGCTCGCGCTCCCGTTCCAAGACCAG
Seq C2 exon
GCAAATCCAGACCCAAACTGCTGCCTGGGAGTGTTCGGATTGAGCCTGTACACCACAGAGAGAGACCTGAGGGAGGTCTTCTCTAAATATGGCCCTCTGAGTGATGTGTGCATTGTGTACGACCAGCAGTCACGGCGCTCCAGGGGTTTTGCTTTTGTCTACTTTGAGAACAGAGAAGATTCAAAAGAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSDARG00000002168_MULTIEX2-2/4=C1-C2
Average complexity
C3
Mappability confidence:
100%=100=100%
Protein Impact
ORF disruption upon sequence exclusion
No structure available
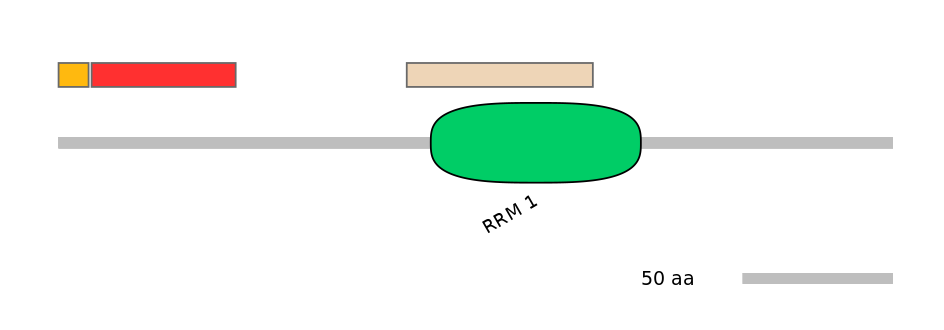
Features
Disorder rate (Iupred):
C1=0.667 A=1.000 C2=0.065
Domain overlap (PFAM):
C1:
PF131361=DUF3984=PU(10.8=90.9),PF086487=DUF1777=PU(8.2=81.8),PF072186=RAP1=PU(8.1=72.7)
A:
PF131361=DUF3984=FE(51.6=100),PF086487=DUF1777=FE(43.6=100),PF072186=RAP1=FE(48.5=100)
C2:
PF044847=DUF566=FE(32.6=100),PF0007617=RRM_1=PU(76.1=85.7)
Main Skipping Isoform:
NA
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Conservation
Mouse
(mm9)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
GTTTGTGTGTGCGTTTGAGCA
R:
CTGCTGGTCGTACACAATGCA
Band lengths:
253-399
Functional annotations
There are 1 annotated functions for this event
PMID: 14709600
Presence of the CLK2 kinase prevents the usage of exons 2 (HsaEX0066781) and 3 (HsaEX0066782), generating the htra2-beta3 mRNA. The resulting HTRA2-BETA3 protein lacks the first RS domain of HTRA2-BETA1, is expressed in several tissues and has no influence on tra2-beta splice site selection. It is proposed that the regulation of the tra2-beta pre-mRNA alternative splicing provides a robust and sensitive molecular sensor that measures the ratio between HTRA2-BETA1 and its interacting proteins.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]