DmeEX0009895 @ dm6
Exon Skipping
Gene
FBgn0003742 | tra2
Description
The gene transformer 2 is referred to in FlyBase by the symbol Dmel ra2 (CG10128, FBgn0003742). It is a protein_coding_gene from Dmel. It has 8 annotated transcripts and 8 polypeptides (4 unique). Gene sequence location is 2R:14602004..14604337. Its molecular function is described by: identical protein binding; RNA binding; mRNA binding; pre-mRNA binding; protein binding. It is involved in the biological process described with 12 unique terms, many of which group under: developmental process; reproduction; developmental process involved in reproduction; cellular component organization or biogenesis; male somatic sex determination. 70 alleles are reported. The phenotypes of these alleles manifest in: abdominal sternite 6; female genital disc; segment of leg; female genitalia; adult prothoracic segment. The phenotypic classes of alleles include: phenotype; fertile; visible; sterile. Summary of modENCODE Temporal Expression Profile: Temporal profile ranges from a peak of moderately high expression to a trough of moderate expression. Peak expression observed within 00-12 hour embryonic stages, during early pupal stages.
Coordinates
chr2R:14603348-14604337:-
Coord C1 exon
chr2R:14604197-14604337
Coord A exon
chr2R:14603984-14604123
Coord C2 exon
chr2R:14603348-14603501
Length
140 bp
Sequences
Splice sites
3' ss Seq
CATTGCTCCGATAAGCGAAGTTT
3' ss Score
-5.34
5' ss Seq
CAGGTAAAT
5' ss Score
8.76
Exon sequences
Seq C1 exon
TCCTCACCTATCGCCACCCCTGTTGAAGCGTGCGTCTTTCTAAACGCAGACAGAATAAATAAAACCTGCCAAAGGCTATCAATTAATTGGTAAACACTGATTGGGTGTGCAATATAGCAGGGAATCAAACCCTCAAAAATG
Seq A exon
TTTCGATAGGTAAGCAAAAAGCCAATGGATCGGGAGCCACTCAGTAGCGGGCGTCTCCATTGCAGCGCCAGATATAAACACAAGCGATCGGCGTCATCATCGTCAGCGGGGACAACTTCATCCGGACACAAGGACCGCAG
Seq C2 exon
GTCTGACTACGATTACTGTGGCAGTCGCCGGCACCAGCGGTCCTCCTCTCGCCGACGCTCCCGTTCGCGTTCCTCCTCGGAGTCGCCGCCACCGGAGCCGCGTCATCGCTCCGGACGTTCATCACGCGATCGCGAACGAATGCACAAGTCTCGC
VastDB Features
Vast-tools module Information
Secondary ID
FBgn0003742-'0-10,'0-2,2-10
Average complexity
C1
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (Ref, Alt. ATG (>10 exons))
No structure available
Features
Disorder rate (Iupred):
C1=NA A=1.000 C2=0.987
Domain overlap (PFAM):
C1:
NA
A:
NO
C2:
NO
Main Inclusion Isoform:
FBpp0088562
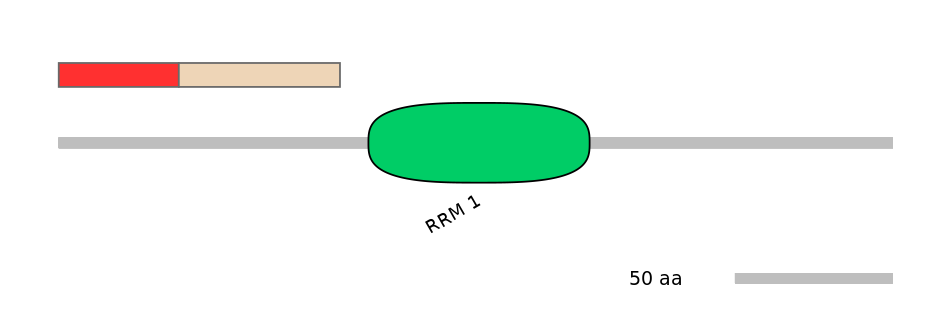
Main Skipping Isoform:
FBpp0088560
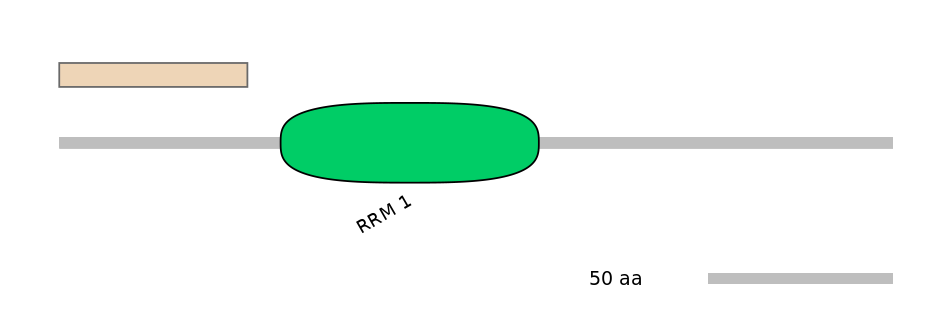
Other Inclusion Isoforms:
FBpp0088563, FBpp0088946, FBpp0088947
Other Skipping Isoforms:
NA
Associated events
Conservation
Primers PCR
Suggestions for RT-PCR validation
F:
TGTTGAAGCGTGCGTCTTTCT
R:
GATGAACGTCCGGAGCGATG
Band lengths:
244-384
Functional annotations
There are 1 annotated functions for this event
PMID: 14709600
Presence of the CLK2 kinase prevents the usage of exons 2 (HsaEX0066781) and 3 (HsaEX0066782), generating the htra2-beta3 mRNA. The resulting HTRA2-BETA3 protein lacks the first RS domain of HTRA2-BETA1, is expressed in several tissues and has no influence on tra2-beta splice site selection. It is proposed that the regulation of the tra2-beta pre-mRNA alternative splicing provides a robust and sensitive molecular sensor that measures the ratio between HTRA2-BETA1 and its interacting proteins.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Neural diversity
- Neurogenesis
- Neuronal activity
- Splicing factor regulation (brain)
- Splicing factor regulation (SL2)