GgaEX6018560 @ galGal4
Exon Skipping
Gene
ENSGALG00000007260 | ZEB1
Description
zinc finger E-box-binding homeobox 1 [Source:RefSeq peptide;Acc:NP_990462]
Coordinates
chr2:14310394-14315784:-
Coord C1 exon
chr2:14315679-14315784
Coord A exon
chr2:14312446-14314256
Coord C2 exon
chr2:14310394-14310574
Length
1811 bp
Sequences
Splice sites
3' ss Seq
TTTCTTTTTTTAATCTTTAGGAG
3' ss Score
10.41
5' ss Seq
CAGGTACCG
5' ss Score
10.05
Exon sequences
Seq C1 exon
AGACATGTGACGCAGTCCAGTGGTAATCGAAAATTCAAGTGCACTGAATGTGGAAAAGCTTTCAAATATAAACATCATCTAAAGGAACACCTACGAATCCACAGTG
Seq A exon
GAGAGAAGCCATATGAGTGCCCAAACTGCAAGAAACGTTTTTCCCATTCTGGTTCATACAGCTCACACATAAGCAGTAAGAAGTGTATTGGTTTGATGCCCGTGAATGGTCGAGCTCGGTCAGGGCTCAAGATGTCTCAGTGCTCCTCCCCTTCCCTTTCTGCATCGCCCGGTAGCCCAGCAAGACCACAGATACGACAAAAGATAGAAAATAAACCCTTGCAAGAGCAACTTCCTGTTAACCAAATTAAAACTGAACCTGTGGATTATGAATTCAAGCCCATAGTGGTTGCTTCAGGAATTAATTGTTCGACCCCTTTGCAGAATGGGGTTTTTAGTGGTGGTAGCCCGTTGCAGGCAACCAGTTCTCCTCAGGGTGTGGTGCAAGCTGTTGTTCTGCCAACAGTGGGTCTGGTGTCTCCCATAAGCATCAACTTAAGTGACATTCAAAATGTACTTAAAGTGGCAGTGGATGGTAATGTAATAAGGCAAGTATTGGAAAACAATCACGCTAATCTTGCATCCAAAGAACAAGAAACAATCAGCAATGCATCTATACAACAAGCTGGCCATTCCCTCATTTCAGCTATCAGTCTTCCTTTGGTTGACCAAGATGGGACAACCAAAATTATCATCAACTACAGCTTGGAGCAGCCAAGTCAACTTCAGGTTGTTCCCCAAAATCTAAAAAAAGAACATTCTGTTCCTACAAACAGTTGCAAAAATGAAAAGTTACCAGAAGATCTTACGGTGAAGTCTGAGAAAGATAAAAACTTTGAAGGAGAGACCAACGATAGCACTTGTCTTCTTTGTGATGACTGTCCAGGAGATCTTAATGCACTTCAAGAATTAAAGCACTATGAAACAAAAAATCCTCCTCAGCTTCCCCAGTCCAGTGGAACAGAAGCTGAGAAGCCCAGCTCCCCTGCCCCATCAGAAACTGGGGAGAACAACTTATCTCCTGGTCAGCCACCCTTAAAGAACCTTTTATCGCTCCTAAAAGCATATTATGCATTAAATGCACAACCAAGCGCAGAAGAGCTTTCAAAAATTGCCGATTCCGTGAACCTACCACTGGATGTGGTAAAAAAGTGGTTTGAAAAAATGCAAGCTGGACAAATTTCTGTGCAGTCTTCTGGACCATCTTCTCCTGAACAAGTTAAAATAAGCAGTCCCACAGATAACGATGATCAAGCAGCAACTACAAATGAGAGTGAACCCCAGAACAGCACAAACAACTCACAAAATCCGGCCAATACAAGTAAATCTCAGACTTCATCAGGGGGATCAACTCAGAATGGTTCGCGCAGTAGCACGCCATCCCCATCACCACTAAACCTTTCTTCATCAAGAAATTCACAGGGTTATACGTACACAGCAGAGGGTGTACAAGAAGAGCCACAAATGGAACCTCTTGACCTTTCACTACCAAAGCAACATGGAGAACTGTTGGAAAGATCTACCATAACTAGTGTTTACCAGAACAGTGTTTATTCTGTCCAGGAAGAACCTTTGAACTTAACTTGTGCAAAAAAAGAACCACAAAAGGACAACAGTATTACAGACTCTGATCCTATTGTAAATGTAATCCCACCAAGTGCCAATCCCATAAATATTGCTATACCTACAGTCACTGCCCAGTTACCTACAATTGTTGCCATTGCTGACCAGAACAGTGTTCCATGCTTGAGAGCTCTTGCTGCCAATAAGCAAACCATTTTGATTCCCCAGGTGGCTTATACGTACTCTACTACAGTTAGTCCTGCAGTTCAGGAAACACCACCAAAACAGACCCAAGCCAACGGAAGTCAG
Seq C2 exon
GACGAAAGGCAGGACACTAGCTCAGAAGGAGTATCGAATGTAGAAGATCAAAATGATTCTGATTCAACACCCCCAAAGAAAAAGATGAGAAAGACAGAAAATGGGATGTATGCATGTGATTTGTGTGACAAAATATTCCAGAAGAGCAGCTCATTATTGAGACATAAGTATGAACACACAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSGALG00000007260-'20-20,'20-18,22-20=AN
Average complexity
A_S
Mappability confidence:
100%=100=100%
Protein Impact
ORF disruption upon sequence exclusion
No structure available
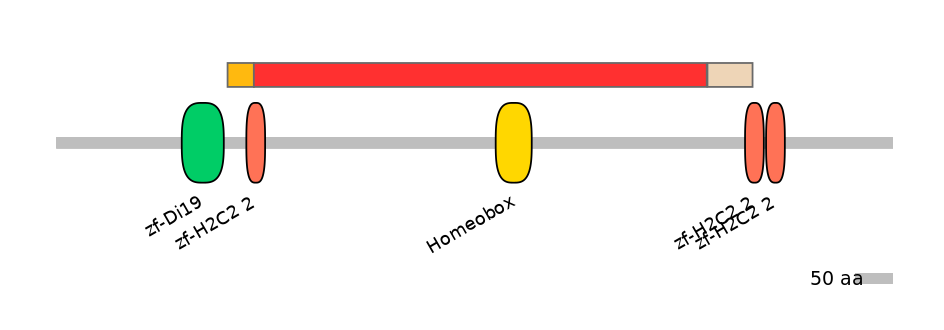
Features
Disorder rate (Iupred):
C1=0.167 A=0.563 C2=0.623
Domain overlap (PFAM):
C1:
PF134651=zf-H2C2_2=PU(38.5=27.8)
A:
PF134651=zf-H2C2_2=PD(57.7=2.5),PF0004624=Homeobox=WD(100=8.1)
C2:
PF134651=zf-H2C2_2=PU(38.5=16.4)
Main Skipping Isoform:
NA
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Other assemblies
Conservation
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
ACGCAGTCCAGTGGTAATCGA
R:
CTGTGTGTTCATACTTATGTCTCAA
Band lengths:
278-2089
Functional annotations
There are 1 annotated functions for this event
PMID: 10567582
Encodes an experimentally validated Eukaryotic Linear Motif (ELM). Method: two hybrid. ELM ID: ELMI000085; ELM sequence: PLDLS; Overlap: FULL
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]