HsaEX0033409 @ hg19
Exon Skipping
Gene
ENSG00000088247 | KHSRP
Description
KH-type splicing regulatory protein [Source:HGNC Symbol;Acc:6316]
Coordinates
chr19:6419214-6420482:-
Coord C1 exon
chr19:6420433-6420482
Coord A exon
chr19:6420084-6420155
Coord C2 exon
chr19:6419214-6419271
Length
72 bp
Sequences
Splice sites
3' ss Seq
CTCCTTCTTTCTCCATCCAGTCA
3' ss Score
9.14
5' ss Seq
CAGGTATAA
5' ss Score
7.46
Exon sequences
Seq C1 exon
GACTTCAATGACAGAAGAGTACAGGGTCCCAGACGGCATGGTGGGCCTGA
Seq A exon
TCATTGGCAGAGGAGGTGAACAAATTAACAAAATCCAACAGGATTCAGGCTGCAAAGTACAGATTTCTCCAG
Seq C2 exon
ACAGCGGTGGCCTACCCGAGCGCAGTGTGTCCTTGACAGGAGCCCCAGAATCTGTCCA
VastDB Features
Vast-tools module Information
Secondary ID
ENSG00000088247_CASSETTE1
Average complexity
S
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (Ref)
No structure available
Features
Disorder rate (Iupred):
C1=0.833 A=0.800 C2=0.950
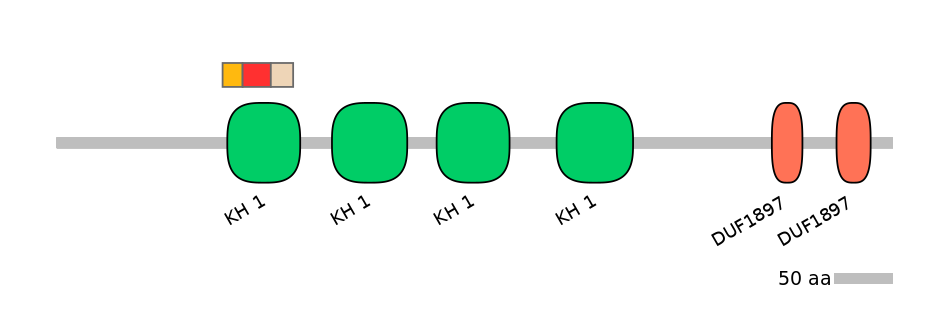
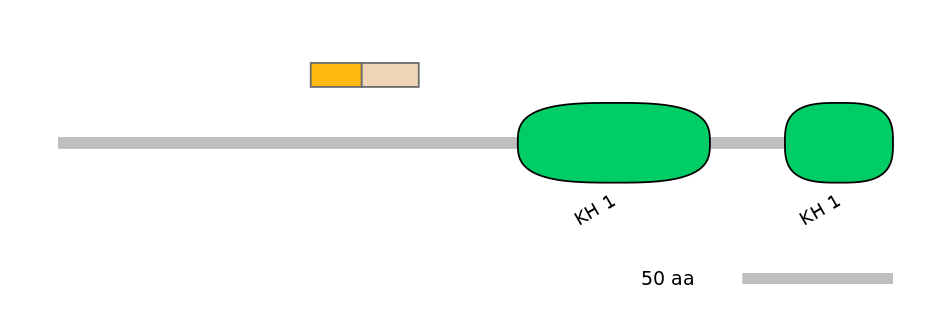
Domain overlap (PFAM):
C1:
PF0001324=KH_1=PU(20.6=72.2)
A:
PF0001324=KH_1=FE(38.1=100)
C2:
PF0001324=KH_1=FE(30.2=100)
Other Skipping Isoforms:
NA
Associated events
Other assemblies
Conservation
Rat
(rn6)
No conservation detected
Chicken
(galGal3)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
CAATGACAGAAGAGTACAGGGTCC
R:
TGGACAGATTCTGGGGCTCC
Band lengths:
103-175
Functional annotations
There are 1 annotated functions for this event
PMID: 19770516
This event
Encodes an experimentally validated Eukaryotic Linear Motif (ELM). Method: coimmunoprecipitation, mutation analysis. ELM ID: ELMI002009; ELM sequence: VQISPD; Overlap: FULL
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Autistic and control brains
- Pre-implantation embryo development
Other AS DBs:
FasterDB (Includes CLIP-seq data)
AS-ALPS (AS-induced ALteration of Protein Structure, links to PINs)
APPRIS (Selection of principal isoform)
DEU primates (Only for human)