HsaEX0043381 @ hg19
Exon Skipping
Gene
ENSG00000163072 | NOSTRIN
Description
nitric oxide synthase trafficker [Source:HGNC Symbol;Acc:20203]
Coordinates
chr2:169658898-169681227:+
Coord C1 exon
chr2:169658898-169659183
Coord A exon
chr2:169668077-169668162
Coord C2 exon
chr2:169681144-169681227
Length
86 bp
Sequences
Splice sites
3' ss Seq
TTATTGTTCTTGTTTTTCAGTAT
3' ss Score
9.89
5' ss Seq
AAGGTACAA
5' ss Score
6.82
Exon sequences
Seq C1 exon
GATCCTGCCCTCTGAGGGTTTACAGAAGTCATTCTCACTATCTCACACTGGCCGTCTTGACCCAGAGCAGTTCTTTACTTTCTCGGGACACAAACGGGATGAAAGTCCAGGGCTGGGACTTCCTCCTTGGAATGTTTGACAAGGACTGACGTACTCTAGGCCCTTTGAATCCCGGAAGTCCAGGACACACCCAACCTTGATTACAGTGTCCAGAATGCTGGAGCGTAGGTGAAAGGACAAAAGCCAGACACATTTCAACATGAGGGACCCACTGACAGATTGTCCG
Seq A exon
TATAATAAAGTATACAAGAACCTAAAGGAGTTTTCTCAAAATGGAGAGAATTTCTGCAAACAGGTCACATCTGTTCTTCAGCAAAG
Seq C2 exon
GGCAAACCTGGAAATTAGCTATGCCAAAGGACTTCAGAAACTGGCAAGCAAGCTGAGCAAAGCATTACAGAACACGAGAAAAAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSG00000163072_MULTIEX1-5/7=4-7
Average complexity
S
Mappability confidence:
100%=100=100%
Protein Impact
ORF disruption upon sequence exclusion
No structure available
Features
Disorder rate (Iupred):
C1=0.000 A=0.000 C2=0.000
Domain overlap (PFAM):
C1:
PF0061118=FCH=PU(2.3=22.2)
A:
PF0061118=FCH=FE(32.2=100)
C2:
PF0061118=FCH=FE(32.2=100)
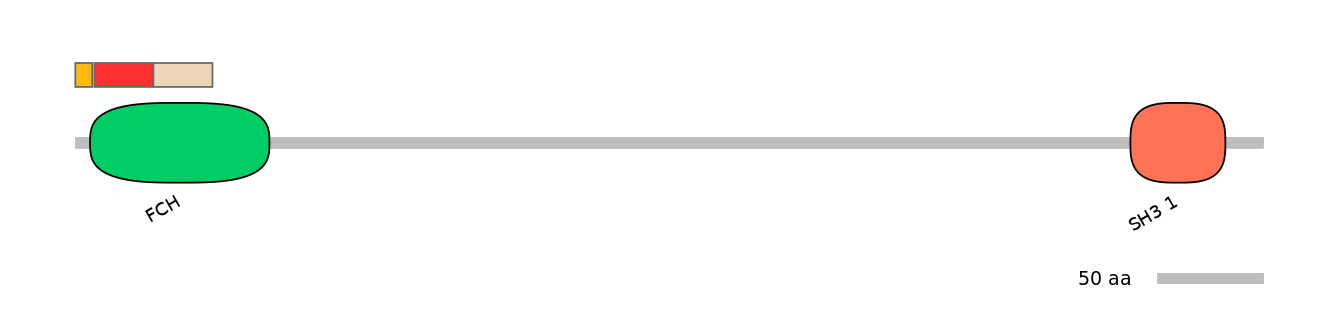
Main Skipping Isoform:
NA
Other Inclusion Isoforms:
Other Skipping Isoforms:
NA
Associated events
Other assemblies
Conservation
Chicken
(galGal4)
No conservation detected
Chicken
(galGal3)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
TTGAATCCCGGAAGTCCAGGA
R:
AGCTTGCTTGCCAGTTTCTGA
Band lengths:
176-262
Functional annotations
There are 2 annotated functions for this event
PMID: 17570224
This event
Exogenously expressed NOSTRINalpha is distributed over the cytosol and associated with subcellular membranes, whereas NOSTRINbeta (skipping of HsaEX0043381) is localized predominantly to the nucleus. Experiments in CHO cells.
PMID: 18980613
This event
This study shows that nuclear import of NOSTRINbeta (skipping of CDS exon HsaEX0043381, inducing the use of a downstream ATG) depends on two nuclear localization signals (aa 32-36: KKRK and aa 57-61: KAKKK). Each of the sequences is independently functional, but both are required to sustain nuclear localization of NOSTRINbeta. Export of NOSTRINbeta from the nucleus is facilitated by a CRM1-dependent mechanism relying on the nuclear export sequence LELEKERIQL (aa 135-145). Unlike NOSTRINbeta, the full-length variant NOSTRINalpha was conspicuously absent from the nucleus. This is most likely because of the fact that its N-terminal F-BAR domain, which is truncated in NOSTRINbeta, facilitates association with cellular membranes. NOSTRINbeta directly binds to the 5'-regulatory region of the NOSTRIN gene (bp -200 to -1), and overexpression of NOSTRINbeta strongly decreases transcription of a reporter gene under control of this DNA region.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Autistic and control brains
- Pre-implantation embryo development
Other AS DBs:
FasterDB (Includes CLIP-seq data)
AS-ALPS (AS-induced ALteration of Protein Structure, links to PINs)
APPRIS (Selection of principal isoform)
DEU primates (Only for human)