MmuEX6101842 @ mm10
Exon Skipping
Gene
ENSMUSG00000034738 | Nostrin
Description
nitric oxide synthase trafficker [Source:MGI Symbol;Acc:MGI:3606242]
Coordinates
chr2:69135800-69152418:+
Coord C1 exon
chr2:69135800-69135916
Coord A exon
chr2:69144770-69144855
Coord C2 exon
chr2:69152335-69152418
Length
86 bp
Sequences
Splice sites
3' ss Seq
TGTTATTTTTTTTCCCACAGTAC
3' ss Score
10.21
5' ss Seq
AAGGTACCA
5' ss Score
7.87
Exon sequences
Seq C1 exon
GAAGTCCAGAACACACCCAACCTGATTGATTGCAGCGTCCAGCATGCTGGATCGAGCGTAGGTAAAAGAAAAAGCAGACAGCATTTCAACATGAGGGACCCACTGACGGACTGTTCG
Seq A exon
TACAATAAAGTATACAAGAGCCTGAAAGAGTTTGCCCAACATGGAGACAATTTCTGCAAGCAGATCACATCTGTTCTTCAGCAAAG
Seq C2 exon
GGCCAACCTGGAGATTAGCTACGCCAAAGGGCTTCAGAAACTGGCCGTCAGGCTCAGCAAAGCACTACAGAGCACAAAGAAAAA
VastDB Features
Vast-tools module Information
Secondary ID
ENSMUSG00000034738-'1-4,'1-2,4-4
Average complexity
S
Mappability confidence:
100%=100=100%
Protein Impact
ORF disruption upon sequence exclusion
No structure available
Features
Disorder rate (Iupred):
C1=0.000 A=0.000 C2=0.000
Domain overlap (PFAM):
C1:
PF050557=DUF677=PU(5.9=44.4),PF0061118=FCH=PU(4.5=33.3)
A:
PF050557=DUF677=FE(41.2=100),PF0061118=FCH=FE(41.8=100)
C2:
PF050557=DUF677=FE(41.2=100),PF0061118=FCH=FE(41.8=100)
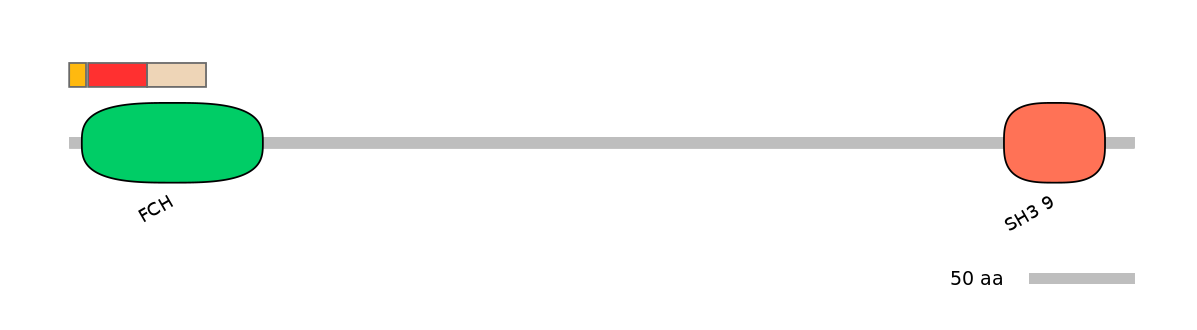
Main Skipping Isoform:
ENSMUST00000041865fB13709

Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Other assemblies
Conservation
Chicken
(galGal4)
No conservation detected
Chicken
(galGal3)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
GTCCAGAACACACCCAACCTG
R:
TGTAGTGCTTTGCTGAGCCTG
Band lengths:
183-269
Functional annotations
There are 2 annotated functions for this event
PMID: 17570224
Exogenously expressed NOSTRINalpha is distributed over the cytosol and associated with subcellular membranes, whereas NOSTRINbeta (skipping of HsaEX0043381) is localized predominantly to the nucleus. Experiments in CHO cells.
PMID: 18980613
This study shows that nuclear import of NOSTRINbeta (skipping of CDS exon HsaEX0043381, inducing the use of a downstream ATG) depends on two nuclear localization signals (aa 32-36: KKRK and aa 57-61: KAKKK). Each of the sequences is independently functional, but both are required to sustain nuclear localization of NOSTRINbeta. Export of NOSTRINbeta from the nucleus is facilitated by a CRM1-dependent mechanism relying on the nuclear export sequence LELEKERIQL (aa 135-145). Unlike NOSTRINbeta, the full-length variant NOSTRINalpha was conspicuously absent from the nucleus. This is most likely because of the fact that its N-terminal F-BAR domain, which is truncated in NOSTRINbeta, facilitates association with cellular membranes. NOSTRINbeta directly binds to the 5'-regulatory region of the NOSTRIN gene (bp -200 to -1), and overexpression of NOSTRINbeta strongly decreases transcription of a reporter gene under control of this DNA region.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Ribosome-engaged transcriptomes of neuronal types
- Neural differentiation time course
- Muscular differentiation time course
- Spermatogenesis cell types
- Reprogramming of fibroblasts to iPSCs
- Hematopoietic precursors and cell types