HsaEX0045711 @ hg38
Exon Skipping
Gene
ENSG00000007372 | PAX6
Description
paired box 6 [Source:HGNC Symbol;Acc:HGNC:8620]
Coordinates
chr11:31784792-31793553:-
Coord C1 exon
chr11:31793438-31793553
Coord A exon
chr11:31790710-31790860
Coord C2 exon
chr11:31784792-31790019
Length
151 bp
Sequences
Splice sites
3' ss Seq
GTTTGCCTCTCTCCTCACAGCCC
3' ss Score
11.45
5' ss Seq
CAGGTGAGC
5' ss Score
9.6
Exon sequences
Seq C1 exon
TTTCCTCCTTCACATCTGGCTCCATGTTGGGCCGAACAGACACAGCCCTCACAAACACCTACAGCGCTCTGCCGCCTATGCCCAGCTTCACCATGGCAAATAACCTGCCTATGCAA
Seq A exon
CCCCCAGTCCCCAGCCAGACCTCCTCATACTCCTGCATGCTGCCCACCAGCCCTTCGGTGAATGGGCGGAGTTATGATACCTACACCCCCCCACATATGCAGACACACATGAACAGTCAGCCAATGGGCACCTCGGGCACCACTTCAACAG
Seq C2 exon
GACTCATTTCCCCTGGTGTGTCAGTTCCAGTTCAAGTTCCCGGAAGTGAACCTGATATGTCTCAATACTGGCCAAGATTACAGTAAAAAAAAAAAAAAAAAAAAAAAGGAAAGGAAATATTGTGTTAATTCAGTCAGTGACTATGGGGACACAACAGTTGAGCTTTCAGGAAAGAAAGAAAAATGGCTGTTAGAGCCGCTTCAGTTCTACAATTGTGTCCTGTATTGTACCACTGGGGAAGGAATGGACTTGAAACAAGGACCTTTGTATACAGAAGGCACGATATCAGTTGGAACAAATCTTCATTTTGGTATCCAAACTTTTATTCATTTTGGTGTATTATTTGTAAATGGGCATTTGTATGTTATAATGAAAAAAAGAACAATGTAGACTGGATGGATGTTTGATCTGTGTTGGTCATGAAGTTGTTTTTTTTTTTTTTAAAAAGAAAACCATGATCAACAAGCTTTGCCACGAATTTAAGAGTTTTATCAAGATAT
VastDB Features
Vast-tools module Information
Secondary ID
ENSG00000007372_MULTIEX2-2/3=1-C2
Average complexity
S
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (No Ref, Alt. Stop)
No structure available
Features
Disorder rate (disopred):
C1=1.000 A=1.000 C2=0.838
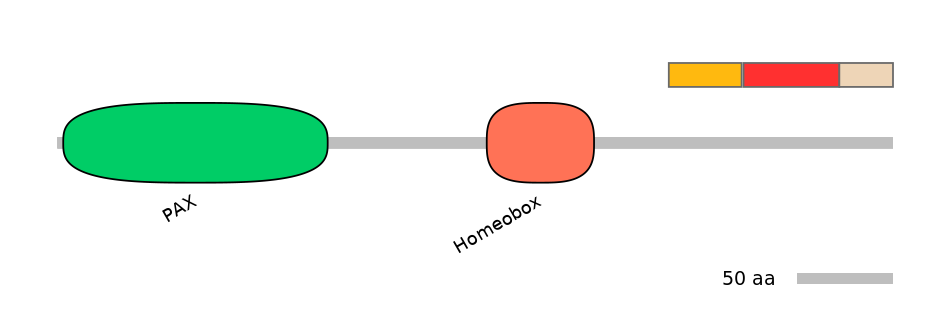
Domain overlap (PFAM):
C1:
NO
A:
NO
C2:
NO
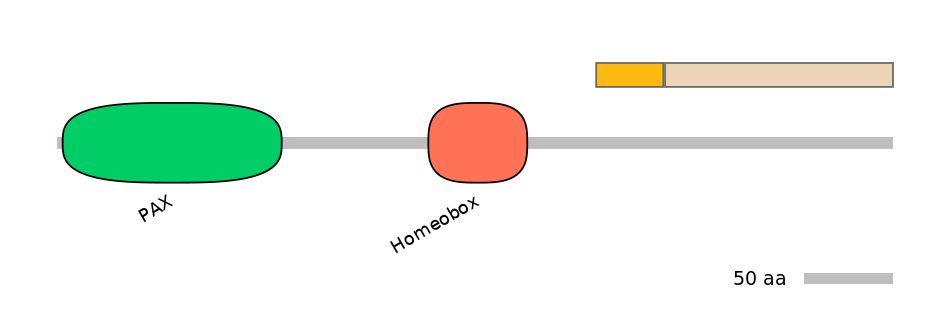
Other Inclusion Isoforms:
ENSP00000241001, ENSP00000368403, ENSP00000368406, ENSP00000368410, ENSP00000368418, ENSP00000368424, ENSP00000368427, ENSP00000388132, ENSP00000404100, ENSP00000404356, ENSP00000431585, ENSP00000480026, ENSP00000490963, ENSP00000490971, ENSP00000491065, ENSP00000491210, ENSP00000491214, ENSP00000491229, ENSP00000491267, ENSP00000491280, ENSP00000491295, ENSP00000491324, ENSP00000491365, ENSP00000491517, ENSP00000491779, ENSP00000491862, ENSP00000491872, ENSP00000491904, ENSP00000491944, ENSP00000492024, ENSP00000492081, ENSP00000492166, ENSP00000492181, ENSP00000492205, ENSP00000492296, ENSP00000492315, ENSP00000492437, ENSP00000492476, ENSP00000492587, ENSP00000492658, ENSP00000492756, ENSP00000492769, ENSP00000492802, ENSP00000492808, ENSP00000492822
Associated events
Other assemblies
Conservation
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
AGACACAGCCCTCACAAACAC
R:
GCTCAACTGTTGTGTCCCCAT
Band lengths:
242-393
Functional annotations
There are 1 annotated functions for this event
PMID: 10329718
Partially encodes an experimentally validated Eukaryotic Linear Motif (ELM). Method: . ELM ID: ELMI001441; ELM sequence: GLISPGV; Overlap: PARTIAL_RIGHT
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Autistic and control brains
- Pre-implantation embryo development