HsaEX0056829 @ hg19
Exon Skipping
Gene
ENSG00000148396 | SEC16A
Description
SEC16 homolog A (S. cerevisiae) [Source:HGNC Symbol;Acc:29006]
Coordinates
chr9:139338275-139340171:-
Coord C1 exon
chr9:139340097-139340171
Coord A exon
chr9:139339504-139339563
Coord C2 exon
chr9:139338275-139338352
Length
60 bp
Sequences
Splice sites
3' ss Seq
GTGTTCTTGCTGTTTTGTAGCTT
3' ss Score
8.88
5' ss Seq
CAGGTACCT
5' ss Score
8.16
Exon sequences
Seq C1 exon
GTTTTAAGCTCTGCAGCGTCACTCCCTGGCTCTGAACTCCCCTCCTCCAGGCCTGAGGGTTCCCAGGGAGGAGAG
Seq A exon
CTTTCGCGCTGTAGTTCAATGAGTTCATTATCACGTGAAGTGAGCCAGCATTTTAATCAG
Seq C2 exon
GCTCCTGGCGACCTCCCTGCTGCAGGGGGCCCTCCCAGCGGGGCCATGCCCTTCTACAACCCTGCTCAGCTGGCACAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSG00000148396_MULTIEX1-2/2=1-C2
Average complexity
C1
Mappability confidence:
100%=100=100%
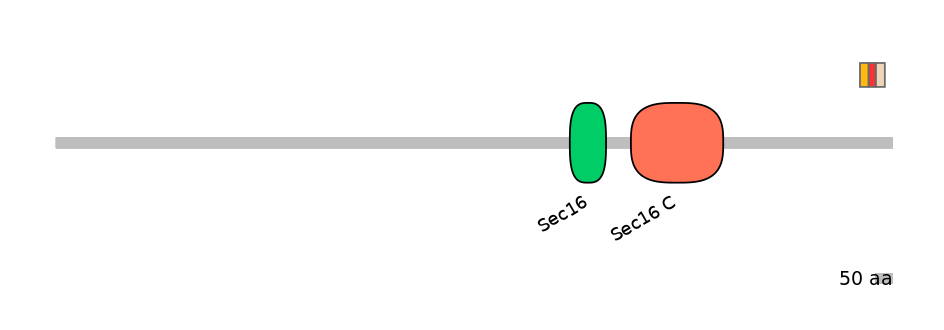
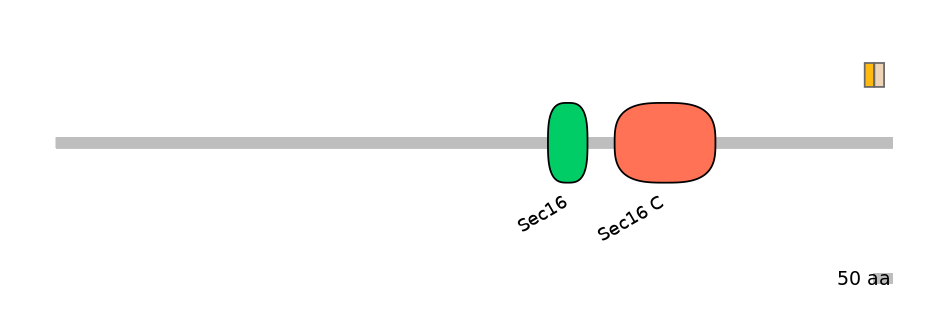
Protein Impact
Alternative protein isoforms (Ref)
No structure available
Features
Disorder rate (Iupred):
C1=1.000 A=1.000 C2=0.992
Domain overlap (PFAM):
C1:
NO
A:
NO
C2:
NO
Associated events
Other assemblies
Conservation
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
CGTCACTCCCTGGCTCTGAA
R:
CCAGCTGAGCAGGGTTGTAGA
Band lengths:
132-192
Functional annotations
There are 1 annotated functions for this event
PMID: 27492621
This event
Activation-induced alternative splicing of Sec16a (inclusion/exclusion of HsaEX0056828 and/or HsaEX0056829) controls adaptation of COPII transport to increased secretory cargo upon T-cell activation. Using splice-site blocking morpholinos and CRISPR/Cas9-mediated genome engineering, the authors show that the number of ER exit sites, COPII dynamics and transport efficiency depend on Sec16 alternative splicing. As the mechanistic basis, the authors suggest the C-terminal Sec16 domain to be a splicing-controlled protein interaction platform, with individual isoforms showing differential abilities to recruit COPII components. These work connects the COPII pathway with alternative splicing, adding a new regulatory layer to protein secretion and its adaptation to changing cellular environments.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- The Cancer Genome Atlas (TCGA)
- Genotype-Tissue Expression Project (GTEx)
- Autistic and control brains
- Pre-implantation embryo development
Other AS DBs:
FasterDB (Includes CLIP-seq data)
AS-ALPS (AS-induced ALteration of Protein Structure, links to PINs)
APPRIS (Selection of principal isoform)
DEU primates (Only for human)