HsaEX0061992 @ hg19
Exon Skipping
Gene
ENSG00000166444 | ST5
Description
suppression of tumorigenicity 5 [Source:HGNC Symbol;Acc:11350]
Coordinates
chr11:8747620-8772272:-
Coord C1 exon
chr11:8772168-8772272
Coord A exon
chr11:8751497-8752756
Coord C2 exon
chr11:8747620-8747756
Length
1260 bp
Sequences
Splice sites
3' ss Seq
TTGTTTTGTTTTGCCCCCAGGTC
3' ss Score
12.18
5' ss Seq
CAGGTAATG
5' ss Score
9.43
Exon sequences
Seq C1 exon
GGCTCTGGGTTTGCAGAGAGCCGAAATGACCATGACTGCCAACAAGAATTCCAGCATCACCCACGGAGCTGGTGGCACTAAAGCCCCTCGGGGGACTCTGAGCAG
Seq A exon
GTCTCAGTCAGTCTCTCCACCTCCAGTTCTCTCCCCACCAAGGAGTCCCATCTACCCGCTCAGTGATAGTGAAACCTCAGCCTGCAGGTACCCCAGCCACTCCAGCTCCCGGGTGCTCCTCAAGGACCGGCACCCCCCAGCTCCTTCACCCCAGAATCCTCAAGATCCCTCCCCAGATACTTCCCCACCCACCTGTCCCTTCAAGACCGCCAGCTTCGGTTATTTGGACAGAAGCCCTTCGGCGTGCAAGAGAGACGCCCAAAAGGAAAGTGTCCAAGGCGCAGCCCAGGATGTAGCAGGGGTCGCTGCCTGCCTCCCCCTTGCCCAGAGCACGCCATTCCCGGGGCCAGCAGCTGGCCCCCGGGGCGTCTTGCTGACCCGTACCGGTACCCGCGCCCACAGCCTGGGCATCCGGGAGAAGATATCAGCATGGGAAGGTCGCCGAGAGGCGTCGCCCAGGATGAGCATGTGTGGAGAGAAGCGGGAGGGCTCTGGGAGCGAGTGGGCGGCCAGTGAGGGCTGCCCCAGCCTGGGCTGTCCCAGCGTGGTGCCGTCCCCCTGCAGCTCTGAAAAGACCTTTGATTTCAAGGGCCTCCGGAGGATGAGCAGGACCTTCTCCGAGTGTTCCTACCCAGAGACTGAGGAGGAGGGAGAGGCGCTCCCTGTCCGGGACTCTTTCTACCGGCTGGAGAAACGGCTGGGCCGGAGTGAGCCCAGCGCCTTCCTCAGGGGGCATGGCAGCAGGAAGGAGAGCTCAGCAGTGCTGAGCCGGATCCAGAAAATTGAACAGGTCCTGAAGGAGCAGCCGGGCCGGGGGCTCCCCCAGCTCCCCAGCAGCTGCTACAGCGTGGACCGGGGGAAAAGGAAGACTGGAACCTTGGGCTCCTTGGAGGAGCCGGCAGGGGGCGCGAGTGTGAGCGCTGGCAGCCGGGCAGTCGGAGTGGCTGGTGTTGCGGGGGAGGCGGGCCCACCCCCAGAGAGGGAAGGCAGTGGTTCCACTAAGCCCGGGACCCCTGGAAATAGCCCTAGCTCCCAGCGGCTGCCATCGAAGAGTTCCCTCGATCCCGCTGTGAACCCTGTCCCCAAACCCAAGCGCACCTTTGAATACGAGGCTGACAAGAACCCCAAGAGTAAGCCCAGTAATGGTCTACCTCCTTCACCCACACCTGCTGCTCCACCTCCCTTGCCCTCCACCCCAGCCCCGCCAGTCACCCGGAGACCCAAGAAGGACATGCGTGGTCACCGCAAGTCCCAGAGCAG
Seq C2 exon
AAAATCCTTTGAGTTTGAGGATGCATCCAGTCTCCAGTCCCTGTACCCCTCTTCTCCCACTGAGAATGGTACTGAGAACCAACCCAAGTTTGGATCCAAAAGCACTTTAGAAGAAAATGCCTATGAAGATATTGTGG
VastDB Features
Vast-tools module Information
Secondary ID
ENSG00000166444-'10-13,'10-12,11-13
Average complexity
S
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (No Ref)
No structure available
Features
Disorder rate (Iupred):
C1=1.000 A=0.812 C2=0.994
Domain overlap (PFAM):
C1:
NO
A:
NO
C2:
NO
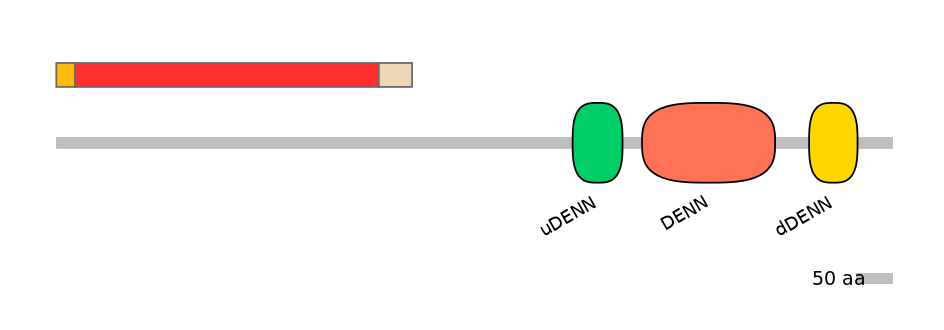
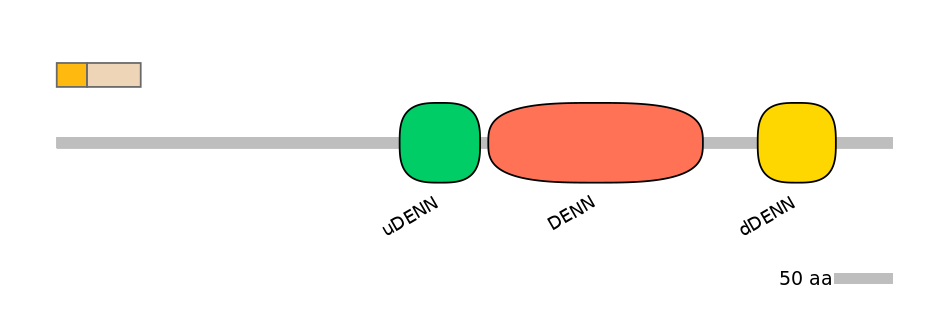
Other Inclusion Isoforms:
ENSP00000319678, ENSP00000350294, ENSP00000431139, ENSP00000431252, ENSP00000431332, ENSP00000431673, ENSP00000431990, ENSP00000432872, ENSP00000433008, ENSP00000433175, ENSP00000433346, ENSP00000433528, ENSP00000433528f10447A, ENSP00000433647, ENSP00000434332, ENSP00000434930, ENSP00000434935, ENSP00000435180, ENSP00000435228, ENSP00000435562, ENSP00000435925, ENSP00000436363, ENSP00000436748
Other Skipping Isoforms:
Associated events
Other assemblies
Conservation
Rat
(rn6)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
GCTCTGGGTTTGCAGAGAGC
R:
CCACAATATCTTCATAGGCATTTTCT
Band lengths:
241-1501
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- The Cancer Genome Atlas (TCGA)
- Genotype-Tissue Expression Project (GTEx)
- Autistic and control brains
- Pre-implantation embryo development
Other AS DBs:
FasterDB (Includes CLIP-seq data)
AS-ALPS (AS-induced ALteration of Protein Structure, links to PINs)
APPRIS (Selection of principal isoform)
DEU primates (Only for human)