HsaEX6016705 @ hg38
Exon Skipping
Gene
ENSG00000077380 | DYNC1I2
Description
dynein cytoplasmic 1 intermediate chain 2 [Source:HGNC Symbol;Acc:HGNC:2964]
Coordinates
chr2:171707287-171715443:+
Coord C1 exon
chr2:171707287-171707377
Coord A exon
chr2:171712767-171712826
Coord C2 exon
chr2:171715328-171715443
Length
60 bp
Sequences
Splice sites
3' ss Seq
GTTGTCCTACAACCATTTAGGAC
3' ss Score
5.51
5' ss Seq
GGGGTATTG
5' ss Score
2.65
Exon sequences
Seq C1 exon
TCCCTCCTCCTATGTCTCCATCCTCCAAATCTGTGAGCACTCCAAGTGAAGCTGGAAGCCAAGACTCTGGAGATGGCGCCGTGGGATCTAG
Seq A exon
GACGCTGCATTGGGATACAGATCCATCAGTTCTTCAGCTTCACTCAGATTCCGATTTGGG
Seq C2 exon
ACGAGGACCTATTAAACTTGGAATGGCTAAAATCACGCAAGTCGACTTTCCTCCTCGAGAAATTGTCACGTATACAAAGGAAACTCAGACTCCAGTTATGGCTCAACCCAAAGAAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSG00000077380-'21-33,'21-32,26-33=AN
Average complexity
A_S
Mappability confidence:
82%=100=100%
Protein Impact
Alternative protein isoforms (Ref)
No structure available
Features
Disorder rate (Iupred):
C1=0.971 A=1.000 C2=0.878
Domain overlap (PFAM):
C1:
PF115403=Dynein_IC2=PU(0.1=0.0)
A:
PF115403=Dynein_IC2=PU(0.1=0.0)
C2:
PF115403=Dynein_IC2=WD(100=82.5)
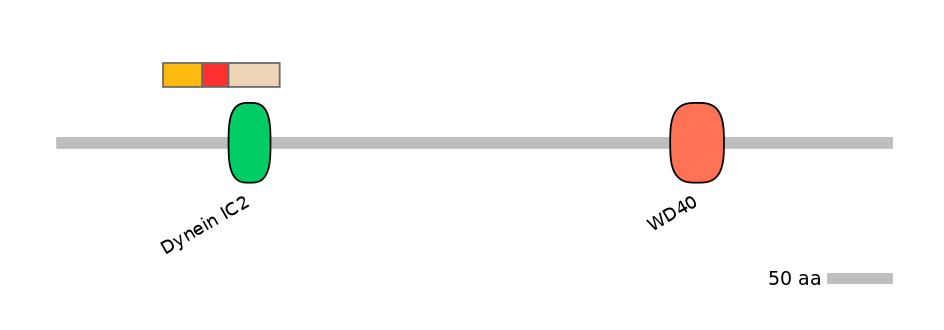
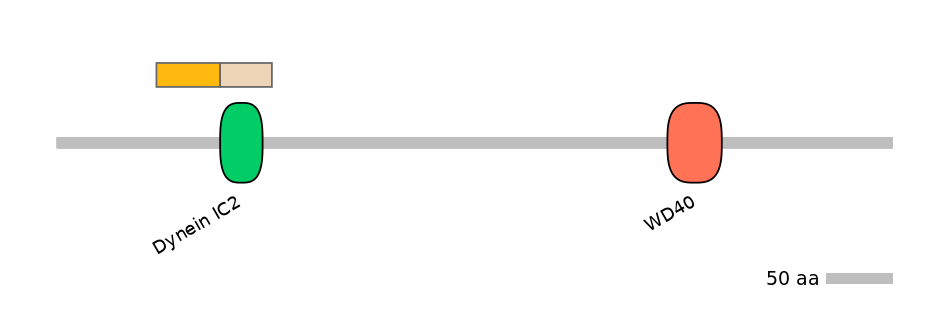
Other Inclusion Isoforms:
Other Skipping Isoforms:
Associated events
Other assemblies
Conservation
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
CCCTCCTCCTATGTCTCCATCC
R:
GAGGAGGAAAGTCGACTTGCG
Band lengths:
146-206
Functional annotations
There are 1 annotated functions for this event
PMID: 22749401
Inclusion decreases interaction with PAFAH1B1 and DYNC1I1, but not with NDEL1, DYNLRB1, DCTN1, DYNLT3, DCTN2, NR3C1. By LUMIER.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Autistic and control brains