HsaEX6054692 @ hg38
Exon Skipping
Gene
ENSG00000183960 | KCNH8
Description
potassium voltage-gated channel subfamily H member 8 [Source:HGNC Symbol;Acc:HGNC:18864]
Coordinates
chr3:19510363-19515428:+
Coord C1 exon
chr3:19510363-19510401
Coord A exon
chr3:19512974-19513325
Coord C2 exon
chr3:19515319-19515428
Length
352 bp
Sequences
Splice sites
3' ss Seq
TTATCCACTGATTTAGTCAGAGC
3' ss Score
-1.98
5' ss Seq
AAGGTAAGA
5' ss Score
10.57
Exon sequences
Seq C1 exon
GTGATATCAAGACTATCAAACAAATCTATGGTCTCACAG
Seq A exon
AGCCCAAGGGAAATGGCAACATCAACAAGCGACTCCCATCCATTGTGGAAGATGAGGAAGAGGAGGAGGAGGGGGAGGAAGAGGAGGCAGTCTCCCTCTCTCCCATCTGCACAAGGGGATCTTCTTCGCGCAACAAGAAGGTTGGAAGCAATAAAGCCTACCTGGGCTTAAGCTTAAAGCAACTGGCCTCGGGAACGGTGCCCTTTCACTCGCCTATCAGAGTCTCCAGGTCAAATTCCCCCAAAACCAAGCAGGAAATTGACCCCCCCAACCATAATAAAAGGAAAGAGAAGAACTTGAAATTGCAACTTTCAACTTTGAATAATGCTGGACCCCCAGACCTCAGTCCAAG
Seq C2 exon
TAGGATTGTTGATGGAATTGAAGATGGAAACAGCAGTGAAGAAAGTCAGACTTTTGATTTTGGCTCTGAACGAATCAGATCAGAGCCCAGAATTTCTCCTCCTCTTGGAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSG00000183960-'33-41,'33-40,34-41
Average complexity
C3
Mappability confidence:
100%=100=100%
Protein Impact
In the CDS, with uncertain impact
No structure available
Features
Disorder rate (Iupred):
C1=0.769 A=0.815 C2=0.973
Domain overlap (PFAM):
C1:
NO
A:
NO
C2:
NO
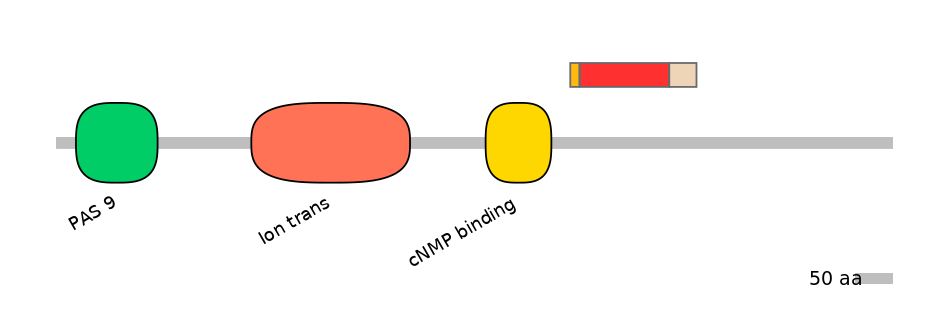
Main Skipping Isoform:
NA
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Other assemblies
Conservation
Rat
(rn6)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
No suggested primer sequences
R:
No suggested primer sequences
Band lengths:
Functional annotations
There are 1 annotated functions for this event
PMID: 27453045
Encodes an experimentally validated Eukaryotic Linear Motif (ELM). Method: alanine scanning, coimmunoprecipitation. ELM ID: ELMI003176; ELM sequence: LPSIVE; Overlap: FULL
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Autistic and control brains