HsaEX6080566 @ hg38
Exon Skipping
Gene
ENSG00000104368 | PLAT
Description
plasminogen activator, tissue type [Source:HGNC Symbol;Acc:HGNC:9051]
Coordinates
chr8:42182719-42189071:-
Coord C1 exon
chr8:42188934-42189071
Coord A exon
chr8:42187906-42188016
Coord C2 exon
chr8:42182719-42182890
Length
111 bp
Sequences
Splice sites
3' ss Seq
TCCTTTTTTATACTTGACAGGTT
3' ss Score
8.73
5' ss Seq
TAGGTGAGT
5' ss Score
8.83
Exon sequences
Seq C1 exon
TGATCTGCAGAGATGAAAAAACGCAGATGATATACCAGCAACATCAGTCATGGCTGCGCCCTGTGCTCAGAAGCAACCGGGTGGAATATTGCTGGTGCAACAGTGGCAGGGCACAGTGCCACTCAGTGCCTGTCAAAA
Seq A exon
GTTGCAGCGAGCCAAGGTGTTTCAACGGGGGCACCTGCCAGCAGGCCCTGTACTTCTCAGATTTCGTGTGCCAGTGCCCCGAAGGATTTGCTGGGAAGTGCTGTGAAATAG
Seq C2 exon
GAAACAGTGACTGCTACTTTGGGAATGGGTCAGCCTACCGTGGCACGCACAGCCTCACCGAGTCGGGTGCCTCCTGCCTCCCGTGGAATTCCATGATCCTGATAGGCAAGGTTTACACAGCACAGAACCCCAGTGCCCAGGCACTGGGCCTGGGCAAACATAATTACTGCCG
VastDB Features
Vast-tools module Information
Secondary ID
ENSG00000104368_MULTIEX1-2/4=1-C2
Average complexity
C2
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (Ref)
No structure available
Features
Disorder rate (Iupred):
C1=0.000 A=0.000 C2=0.000
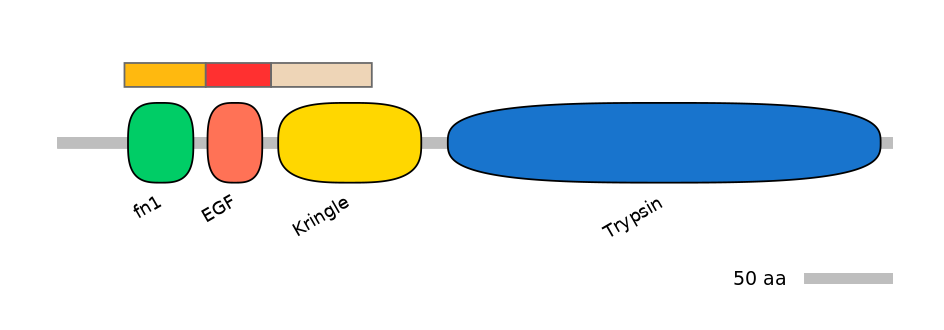
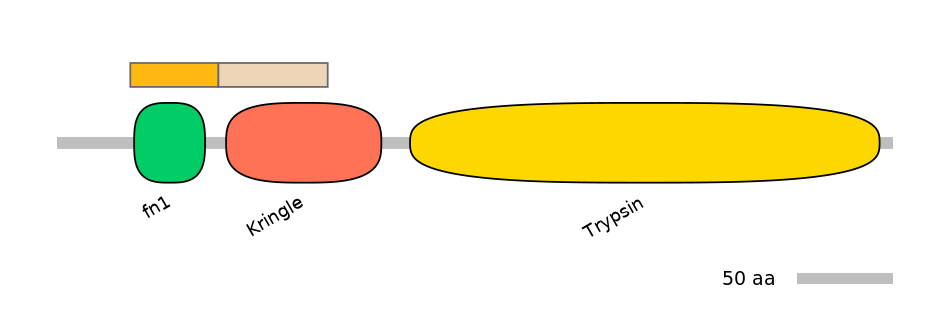
Domain overlap (PFAM):
C1:
PF0003913=fn1=WD(100=80.9)
A:
PF0000822=EGF=WD(100=84.2)
C2:
PF0005113=Kringle=PU(64.6=91.4)
Other Inclusion Isoforms:
Other Skipping Isoforms:
NA
Associated events
Other assemblies
Conservation
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
CCAGCAACATCAGTCATGGCT
R:
CTGGGCACTGGGGTTCTGT
Band lengths:
244-355
Functional annotations
There are 1 annotated functions for this event
PMID: 1900431
Encodes an experimentally validated Eukaryotic Linear Motif (ELM). Method: de novo protein sequencing by mass spectrometry. ELM ID: ELMI000225; ELM sequence: CFNGGTC; Overlap: FULL
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Autistic and control brains