MmuEX0003636 @ mm10
Exon Skipping
Gene
ENSMUSG00000054808 | Actn4
Description
actinin alpha 4 [Source:MGI Symbol;Acc:MGI:1890773]
Coordinates
chr7:28915213-28918791:-
Coord C1 exon
chr7:28918672-28918791
Coord A exon
chr7:28916160-28916246
Coord C2 exon
chr7:28915213-28915300
Length
87 bp
Sequences
Splice sites
3' ss Seq
TCCCTTTGTTCTGTGTGCAGAGA
3' ss Score
9.07
5' ss Seq
AAGGTAAGA
5' ss Score
10.57
Exon sequences
Seq C1 exon
GGGAGCGTTTGCCTAAGCCAGAGCGGGGGAAGATGCGAGTGCACAAGATCAACAATGTAAACAAAGCCCTGGACTTCATCGCCAGCAAGGGAGTCAAGCTGGTGTCCATCGGGGCAGAAG
Seq A exon
AGATTGTGGATGGCAATGCAAAGATGACCCTGGGAATGATCTGGACCATCATCCTCAGATTTGCCATCCAGGACATCTCTGTGGAAG
Seq C2 exon
AGACCTCTGCTAAGGAAGGACTCCTCCTCTGGTGCCAGAGAAAGACAGCCCCATATAAAAACGTCAACGTCCAGAACTTCCATATCAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSMUSG00000054808-'30-12,'30-9,33-12
Average complexity
S
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (Ref)
No structure available
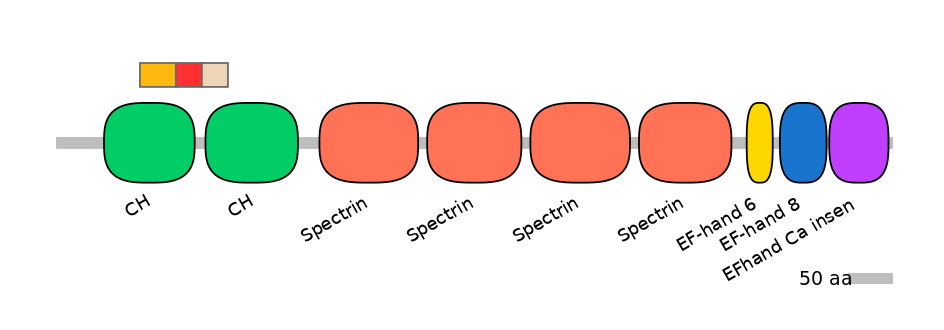
Features
Disorder rate (Iupred):
C1=0.098 A=0.000 C2=0.000
Domain overlap (PFAM):
C1:
PF0030726=CH=FE(39.2=100)
A:
PF0030726=CH=PD(20.6=70.0)
C2:
PF0030726=CH=PU(24.0=83.3)
Main Skipping Isoform:
NA
Other Inclusion Isoforms:
Other Skipping Isoforms:
NA
Associated events
Other assemblies
Conservation
Chicken
(galGal3)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
GAGCGTTTGCCTAAGCCAGAG
R:
GCTGTCTTTCTCTGGCACCAG
Band lengths:
167-254
Functional annotations
There are 3 annotated functions for this event
PMID: 16980305
This study isolated a novel splice variant of alpha-actinin 4 (skipping exons 3-12; ENST00000424234) that is predominantly localized in the nucleus, a pattern distinct from the full-length alpha-actinin 4, which is primarily distributed in the cytoplasm and plasma membrane. Exon 3: HsaEX6094930, ex 4: HsaEX6094929, ex 5: HsaEX6094928, ex 6: HsaEX6094927, ex 7: HsaEX6094926, ex 8: HsaEX0002154, ex 9: HsaEX7010513, ex 10: HsaEX1002558, ex 11: HsaEX6094925, ex 12: HsaEX6094924.
PMID: 22908231
ACTN4(Iso) overexpression increases transcriptional activity of the VDRE and ERE promoters. Both ACTN4(WT) and ACTN4(Iso) co-immunoprecipitate with estrogen receptors. Overexpression of ACTN4(Iso) fused to Gal4 increases basal transcription. The same is seen with overexpression of ACTN4(WT)-Gal4, but the effect is smaller.
PMID: 27998979
Overexpression of either ATN4(WT) or ACTN4(Iso) increases GRE reporter activity. Disruption of the LXXLL nuclear receptor-interacting motif present in ACTN4 results in reduced GR interaction and dexamethasone-mediated transactivation of a GRE reporter while still maintaining its actin-binding activity. In contrast, an ACTN4 isoform, ACTN4 (Iso), that loses its actin-binding domain is still capable of potentiating a GRE reporter.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Pre-implantation embryo development
- Ribosome-engaged transcriptomes of neuronal types
- Neural differentiation time course
- Muscular differentiation time course
- Spermatogenesis cell types
- Reprogramming of fibroblasts to iPSCs
- Hematopoietic precursors and cell types