MmuEX0004933 @ mm10
Exon Skipping
Gene
ENSMUSG00000069601 | Ank3
Description
ankyrin 3, epithelial [Source:MGI Symbol;Acc:MGI:88026]
Coordinates
chr10:69982116-69994446:+
Coord C1 exon
chr10:69982116-69982197
Coord A exon
chr10:69986083-69988632
Coord C2 exon
chr10:69994372-69994446
Length
2550 bp
Sequences
Splice sites
3' ss Seq
TTTCATTTTAAAAATTTAAGAAA
3' ss Score
3.12
5' ss Seq
AAGGTAACC
5' ss Score
9.24
Exon sequences
Seq C1 exon
GCTGAGAAGGCAGACAGACGCCAGAGCTTTGCCTCCCTAGCTTTACGTAAGCGCTACAGCTACTTGACTGAACCCAGCATGA
Seq A exon
AAACAGTTGAACGGAGTTCAGGAACAGCAAGATCCCTCCCCACCACTTATTCACACAAGCCATTCTTTTCTACAAGACCATACCAGTCCTGGACCACGGCTCCAATCACAGTGCCCGGGCCGGCCAAGTCAGGCTCCTTATCCAGCTCTCCCTCTAACACGCCGTCAGCTTCTCCGTTAAAATCCATATGGTCTGTCTCGACGCCTTCTCCGATCAAATCCACGTTAGGCGCCTCAACTACATCTTCAGTCAAATCCATTAGCGACGTGGCGTCTCCTATTAGATCTTTCCGGACAGTTTCTTCGCCAATAAAAACGGTGGTGTCGCCGTCTCCGTACAATCCCCAGGTTGCCTCTGGTACCCTTGGGAGGGTTCCCACCATCACAGAGGCCACACCCATAAAGGGGCTGGCTCCCAACTCAACTTTCTCCTCTCGAACTTCTCCTGTGACGACCGCAGGGTCTCTGTTGGAGAGGTCCTCCATCACCATGACACCCCCTGCCTCCCCCAAATCAAACATCACTATGTATTCCTCAAGCTTGCCATTTAAGTCCATTATCACATCAGCGACACCACTGATCTCTTCCCCTTTAAAGTCAGTGGTGTCTCCGACCAAGTCTGCAGCTGATGTCATCTCAACAGCTAAAGCTACGATGGCATCGTCTCTCTCCTCCCCCTTAAAGCAGATGTCCGGACATGCAGAGGTAGCGCTAGTCAATGGGTCTGTTTCTCCTCTGAAGTACCCTTCGTCTTCAGCTTTAATTAACGGATGCAAAGCCACTGCCACATTACAGGACAAAATTTCTACAGCCACAAACGCTGTGAGCTCGGTGGTGAGCGCAGCCTCTGACACGGTGGAGAAAGCGCTCTCTACCACGACAGCCATGCCCTTTTCCCCACTCAGGTCGTATGTTTCTGCAGCCCCCTCGGCTTTCCAGTCCCTTAGAACTCCCTCTGCAAGTGCACTGTACACCTCCCTCGGGTCCTCGATAGCTGCTACTACCTCATCTGTAACTTCATCAATAATCACAGTGCCAGTATACTCGGTAGTCAATGTTTTGCCAGAACCAGCACTGAAGAAACTCCCAGACTCTAACTCGTTCACCAAGTCGGCAGCGGCTTTGCTGTCGCCCATTAAAACATTGACTACGGAGACACGCCCTCAGCCCCATTTCAATCGAACTTCATCTCCAGTCAAGTCGTCTCTGTTCCTGGCGTCCTCGGCCCTTAAGCCATCAGTACCATCTTCTTTATCTTCCAGCCAGGAGATCTTAAAGGATGTGGCCGAGATGAAGGAAGATCTCATGAGGATGACTGCCATACTGCAGACAGACGTGCCTGAGGAGAAGCCATTCCAAACCGACCTCCCCAGAGAAGGGAGAATAGACGATGAAGAACCTTTCAAAATCGTTGAGAAAGTGAAGGAAGACTTAGTCAAAGTCAGTGAGATACTCAAAAAGGACGTGTGTGTCGAGAGCAAAGGGCCACCCAAGTCCCCGAAGAGTGACAAAGGACACTCTCCAGAGGATGACTGGACAGAATTTAGTTCTGAGGAAATACGAGAAGCCAGGCAGGCTGCGGCCAGTCACGCCCCGTCCCTGCCTGAGAGAGTACACGGGAAGGCCAACCTCACTAGAGTCATAGACTACCTGACCAATGACATCGGGAGCAGCTCACTGACCAACTTAAAGTACAAGTTTGAAGAGGCTAAGAAGGATGGAGAGGAGAGACAGAAGAGGATTTTAAAGCCAGCCATGGCCTTGCAGGAGCATAAACTCAAAATGCCGCCGGCCTCCATGAGGCCTTCCACCTCTGAGAAGGAACTGTGCAAGATGGCTGACTCTTTTTTTGGAGCAGATGCCATCCTAGAGTCTCCCGATGACTTTTCCCAGCATGACCAAGATAAAAGTCCCTTGTCTGACAGTGGCTTTGAAACCCGCAGTGAGAAAACACCTTCGGCCCCGCAAAGTGCCGAGAGCACAGGCCCTAAGCCGTTATTTCACGAAGTCCCCATCCCCCCTGTCATCACAGAGACAAGAACTGAAGTGGTCCATGTGATCAGGAGCTATGAGCCCTCTTCTGGAGAGATCCCCCAGTCCCAGCCAGAGGACCCCGTGTCTCCCAAGCCTTCCCCTACTTTTATGGAGTTGGAACCAAAACCCACCACCTCTAGCATCAAGGAAAAAGTAAAAGCATTTCAGATGAAAGCCAGTAGTGAGGAAGAGGACCACAGTCGGGTTCTAAGCAAAGGCATGCGTGTCAAAGAAGAGACTCACATCACCACGACCACCAGGATGGTGTATCACTCCCCTCCTGGTGGCGAGTGCGCCTCCGAAAGGATTGAGGAGACCATGTCAGTCCATGACATCATGAAGGCCTTTCAGTCCGGGCGAGACCCTTCCAAAGAGCTGGCAGGGCTGTTTGAACACAAGTCGGCCATGTCTCCAGATGTTGCCAAGTCTGCTGCTGAGACCTCAGCCCAGCACGCAGAGAAGGACAGCCAAATGAAACCCAAACTGGAGCGCATTATAGAAGTGCACATCGAAAAAG
Seq C2 exon
GTCCGCAGAGTCCTTGTGAGCGGACGGATATCAGGATGGCGATAGTAGCCGATCACCTGGGACTTAGTTGGACAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSMUSG00000069601_MULTIEX3-3/4=2-C2
Average complexity
S*
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (No Ref)
No structure available
Features
Disorder rate (Iupred):
C1=0.113 A=0.587 C2=0.103
Domain overlap (PFAM):
C1:
NO
A:
NO
C2:
PF0053117=Death=PU(19.0=61.5)
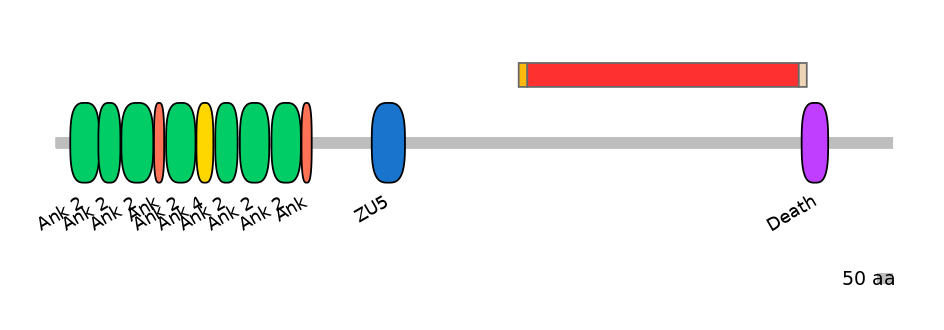
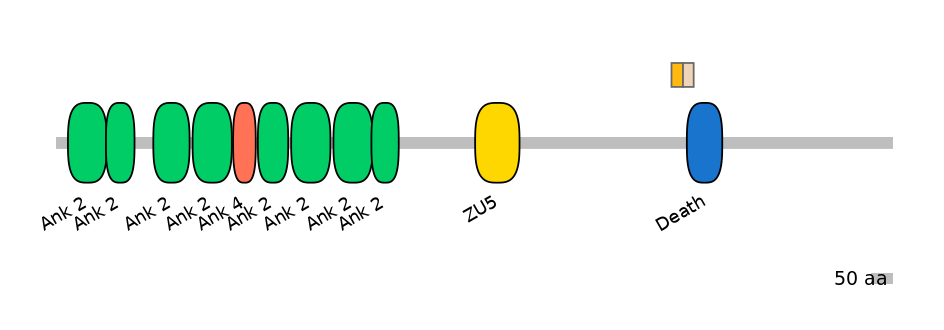
Other Skipping Isoforms:
ENSMUSP00000045834, ENSMUSP00000061698, ENSMUSP00000090087, ENSMUSP00000090088, ENSMUSP00000090089, ENSMUSP00000138095, ENSMUSP00000138123, ENSMUSP00000138285, ENSMUSP00000138326, ENSMUSP00000138337, ENSMUSP00000138347, ENSMUSP00000138348, ENSMUSP00000138356, ENSMUSP00000138413, ENSMUSP00000138481, ENSMUSP00000138531, ENSMUSP00000138586, ENSMUSP00000138623, ENSMUSP00000138671, ENSMUSP00000138770, ENSMUSP00000151906
Associated events
Other assemblies
Conservation
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
No suggested primer sequences
R:
No suggested primer sequences
Band lengths:
Functional annotations
There are 1 annotated functions for this event
PMID: 25552556
This event
Proximal axonal polarity as well as assembly of the AIS and normal morphogenesis of nodes of Ranvier all require a heretofore uncharacterized alternatively spliced giant exon of ankyrin-G (AnkG). This exon has sequence similarity to I-connectin/Titin and was acquired after the first round of whole-genome duplication by the ancestral ANK2/ANK3 gene in early vertebrates before development of myelin. Non-specific knockdown of Ank3 completely abolishes AIS clustering of AnkG, B4 spectrin, VGSC, and NF186. Rescue with 480-kDA isoform (with giant exon; MmuEX0004933) completely restores this phenotype, whereas rescue with the 270-kDa isoform (with a massive ALTD in the giant exon) partially restores the phenotype. Rescue with 190-kDa isoforms fails to restores any part of the phenotype. Specific knockdown of the 480-kDa isoform completely abolishes AnkG staining at the AIS as well as clustering of its associated binding partners, _4 spectrin, VGSC, and NF186.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Pre-implantation embryo development
- Ribosome-engaged transcriptomes of neuronal types
- Neural differentiation time course
- Muscular differentiation time course
- Spermatogenesis cell types
- Reprogramming of fibroblasts to iPSCs
- Hematopoietic precursors and cell types