MmuEX0011716 @ mm9
Exon Skipping
Gene
ENSMUSG00000026034 | Clk1
Description
CDC-like kinase 1 [Source:MGI Symbol;Acc:MGI:107403]
Coordinates
chr1:58476511-58478152:-
Coord C1 exon
chr1:58477927-58478152
Coord A exon
chr1:58476965-58477055
Coord C2 exon
chr1:58476511-58476577
Length
91 bp
Sequences
Splice sites
3' ss Seq
CGCCCACTTGACGTGTCCAGAAG
3' ss Score
4.61
5' ss Seq
GATGTATAG
5' ss Score
-2.49
Exon sequences
Seq C1 exon
CTATTATCTGGAAAGCAGATCCATAAATGAGAAAGCTTATCATAGTCGACGCTATGTTGATGAATACAGGAATGACTACATGGGCTACGAGCCAGGGCATCCCTATGGAGAACCTGGAAGCAGATACCAGATGCATAGTAGCAAGTCCTCTGGTAGGAGTGGAAGAAGCAGTTACAAAAGTAAACACAGGAGTCGCCACCACACTTCGCAGCACCATTCACACGGG
Seq A exon
AAGAGTCACCGAAGGAAAAGATCGAGGAGTGTAGAGGATGATGAGGAGGGTCACCTGATCTGTCAGAGTGGAGACGTACTAAGTGCAAGAT
Seq C2 exon
ATGAAATTGTTGATACTTTAGGTGAAGGTGCTTTCGGAAAAGTGGTGGAATGCATCGATCATAAAGT
VastDB Features
Vast-tools module Information
Secondary ID
ENSMUSG00000026034_CASSETTE2
Average complexity
S
Mappability confidence:
100%=100=100%
Protein Impact
ORF disruption upon sequence exclusion
Show structural model
Features
Disorder rate (Iupred):
C1=0.829 A=0.484 C2=0.000
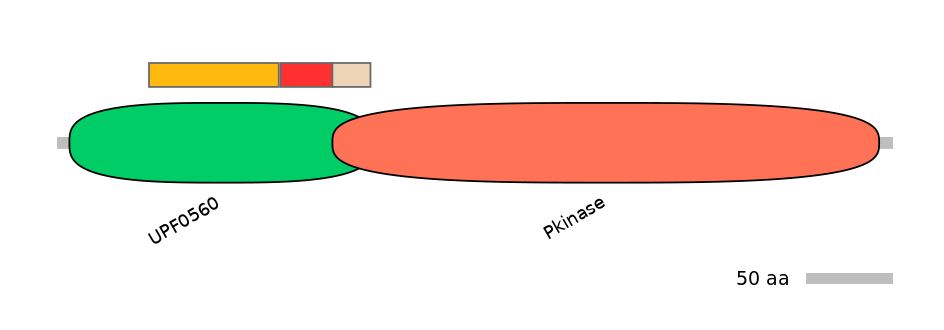
Domain overlap (PFAM):
C1:
PF105774=UPF0560=FE(42.4=100)
A:
PF105774=UPF0560=FE(16.9=100),PF0006920=Pkinase=PU(0.1=0.0)
C2:
PF105774=UPF0560=FE(12.4=100),PF0006920=Pkinase=PU(6.9=95.7)
Main Skipping Isoform:
NA
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Other assemblies
Conservation
Primers PCR
Suggestions for RT-PCR validation
F:
GGGCATCCCTATGGAGAACCT
R:
ACCACTTTTCCGAAAGCACCT
Band lengths:
178-269
Functional annotations
There are 2 annotated functions for this event
PMID: 7665564
Alternative splicing of the primary Sty (another name for CDC-like kinase 1 (CLK1)) transcript generates mRNAs encoding full-length catalytically active (STY) and truncated (via skipping of cassette exon HsaEX0015695), kinase-deficient polypeptides. Both STY and its truncated isoform, STYT, are localized in the nucleus and are capable of heterodimerizing.
PMID: 9315658
The Clk1 kinase, which phosphorylates SR proteins, is regulated through alternative splicing of the Clk1 pre-mRNA, yielding mRNAs encoding catalytically active (includes cassette exon HsaEX0015695) and truncated inactive polypeptides (skips HsaEX0015695) (Clk1 and Clk1T , respectively). The authors present evidence that Clk1 and Clk1T proteins regulate the splicing of Clk1 and adenovirus pre-mRNAs in vivo. The peptide domain encoded by the alternatively spliced exon of Clk1 is essential for the regulatory activity of the Clk1 kinase. This is the first direct demonstration of an in vivo link between alternative splicing and protein kinase activity. (Seems to be a case of auto-regulation of ratio of Clk1 transcripts with/without the cassette exon)
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Pre-implantation embryo development
- Neural differentiation time course
- Muscular differentiation time course
- Spermatogenesis cell types
- Reprogramming of fibroblasts to iPSCs
- Hematopoietic precursors and cell types