DmeEX0004065 @ dm6
Exon Skipping
Gene
FBgn0265998 | Doa
Description
The gene Darkener of apricot is referred to in FlyBase by the symbol DmelDoa (CG42320, FBgn0265998). It is a protein_coding_gene from Dmel. It has 16 annotated transcripts and 16 polypeptides (11 unique). Gene sequence location is 3R:28888073..28922772. Its molecular function is described by: kinase activity; ATP binding; protein kinase activity; protein serine/threonine kinase activity. It is involved in the biological process described with 14 unique terms, many of which group under: multicellular organism development; visual system development; negative regulation of biological process; positive regulation of immune system process; embryo development. 88 alleles are reported. The phenotypes of these alleles manifest in: intracellular organelle; cellular anatomical entity; interommatidial bristle; sex comb; extracellular matrix. The phenotypic classes of alleles include: phenotype; increased mortality; stress response defective; increased mortality during development.
Coordinates
chr3R:28900824-28920291:+
Coord C1 exon
chr3R:28900824-28901022
Coord A exon
chr3R:28919492-28919567
Coord C2 exon
chr3R:28920225-28920291
Length
76 bp
Sequences
Splice sites
3' ss Seq
GATTCGATCGAACCGAACAGACA
3' ss Score
0.54
5' ss Seq
GATGTTCGT
5' ss Score
-5.32
Exon sequences
Seq C1 exon
TTTCGATGACAGCATCTCAACCCGCCGACGCAAGGAGCGTTCGAAGCGTTCGCACCGCAAGTCGCCCGCCGCCAGTCGACGGCAGCACAAATACCGCTACAGGGACGAGACGTCCCATTCGAGCTCCCGCCGGCGGCACCGTGATCGCGCCAAGGACGAACGCGACAGCGGACGCAACAACCGCCAGTCGCAGGCTAAG
Seq A exon
ACAGCAAAGCCCGTCATTCAAGATGATGCTGATGGTCACTTAATTTACCACACCGGAGACATTCTCCATCACAGAT
Seq C2 exon
ATAAGATCATGGCCACACTGGGCGAGGGAACTTTTGGACGTGTGGTCAAGGTCAAAGATATGGAGCG
VastDB Features
Vast-tools module Information
Secondary ID
FBgn0265998_MULTIEX1-6/6=C1-C2
Average complexity
C1
Mappability confidence:
100%=100=100%
Protein Impact
ORF disruption upon sequence exclusion
No structure available
Features
Disorder rate (Iupred):
C1=1.000 A=0.164 C2=0.000
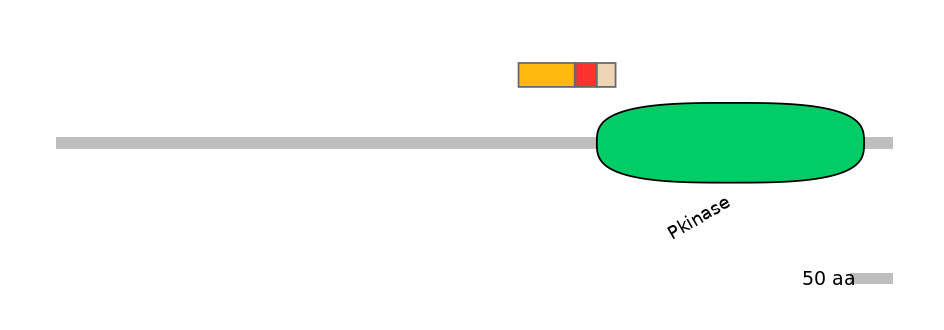
Domain overlap (PFAM):
C1:
NO
A:
PF0006920=Pkinase=PU(0.1=0.0)
C2:
PF0006920=Pkinase=PU(7.0=95.7)
Main Inclusion Isoform:
FBpp0289022
Main Skipping Isoform:
NA
Other Inclusion Isoforms:
FBpp0289020, FBpp0289021, FBpp0289023, FBpp0289025, FBpp0289026, FBpp0289027, FBpp0289028, FBpp0289029, FBpp0289030, FBpp0289031, FBpp0300822, FBpp0300823, FBpp0300824, FBpp0307567
Other Skipping Isoforms:
NA
Associated events
Conservation
Primers PCR
Suggestions for RT-PCR validation
F:
CAGTCGACGGCAGCACAAATA
R:
ACCACACGTCCAAAAGTTCCC
Band lengths:
173-249
Functional annotations
There are 4 annotated functions for this event
PMID: 7665564
Alternative splicing of the primary Sty (another name for CDC-like kinase 1 (CLK1)) transcript generates mRNAs encoding full-length catalytically active (STY) and truncated (via skipping of cassette exon HsaEX0015695), kinase-deficient polypeptides. Both STY and its truncated isoform, STYT, are localized in the nucleus and are capable of heterodimerizing.
PMID: 9315658
The Clk1 kinase, which phosphorylates SR proteins, is regulated through alternative splicing of the Clk1 pre-mRNA, yielding mRNAs encoding catalytically active (includes cassette exon HsaEX0015695) and truncated inactive polypeptides (skips HsaEX0015695) (Clk1 and Clk1T , respectively). The authors present evidence that Clk1 and Clk1T proteins regulate the splicing of Clk1 and adenovirus pre-mRNAs in vivo. The peptide domain encoded by the alternatively spliced exon of Clk1 is essential for the regulatory activity of the Clk1 kinase. This is the first direct demonstration of an in vivo link between alternative splicing and protein kinase activity. (Seems to be a case of auto-regulation of ratio of Clk1 transcripts with/without the cassette exon)
PMID: 16371011
Skipping of exon 4 (HsaEX0015698) increases in brain areas affected by Alzheimer's Disease resulting in increased expression levels of clk2tr. Increased expression of clk2 full-length induces skipping of clk2 exon 4. Increased expression of clk2tr does not promote skipping of clk2 exon 4.
PMID: 31911676
[CRISPR screen]. Conserved poison exon with negative fitness/survivability impact when depleted: HeLa Depleted at 14 days (FC=0.725, FDR=0.000), PC9 No Change at 14 days (FC=1.106, FDR=0.829), Late Xenograft No change (FC=1.369, FDR=0.260).
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Neural diversity
- Neurogenesis
- Neuronal activity
- Splicing factor regulation (brain)
- Splicing factor regulation (SL2)