DreEX0022145 @ danRer10
Exon Skipping
Gene
ENSDARG00000062954 | clk2a
Description
CDC-like kinase 2a [Source:ZFIN;Acc:ZDB-GENE-030131-298]
Coordinates
chr19:9236583-9239747:+
Coord C1 exon
chr19:9236583-9236850
Coord A exon
chr19:9238800-9238890
Coord C2 exon
chr19:9239681-9239747
Length
91 bp
Sequences
Splice sites
3' ss Seq
CTGACCTGTGGCCCAAGCAGCGG
3' ss Score
3.17
5' ss Seq
GATGTACAC
5' ss Score
-3.63
Exon sequences
Seq C1 exon
CTACGACAACCGTTCTTCAGACCGCAGGCCATACGACAGACACTACGGCGAGAGCTACCGGCGCTTGGACCACAGCAGGGAACGGGACAGGGATCAAAACAGGGATCACCGGACGCCGAGCAACTACTACTCTCGCGACGCCTCGCCCAGCTACGACTTTCGCCGGGGACGCGACCGGGACCGCGAGGACTCATACCGGAGGAAAGGCAGCAGGCGTAAGCACAAACGAAGGCGGCGTAGAACCAGGTCCTATAGCGCATCCTCCTCG
Seq A exon
CGGAGTGACAGCGGGACGAGGGCACTGTGTGTGAGGGACGACGAGGAGGGTCACCTGATCTGTCGGAGTGGCGACGTCCTGCAAGAGAGAT
Seq C2 exon
ATGAGATTGTCAGCACTTTGGGTGAGGGCACCTTCGGGAGGGTGATGCAATGTATTGATCATCGCAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSDARG00000062954_CASSETTE1
Average complexity
S*
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (No Ref, Alt. Stop)
No structure available
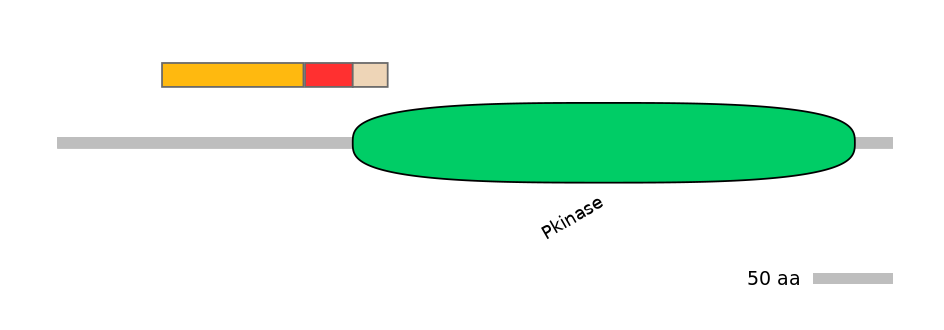
Features
Disorder rate (Iupred):
C1=0.992 A=0.280 C2=0.108
Domain overlap (PFAM):
C1:
NO
A:
PF0006920=Pkinase=PU(0.1=0.0)
C2:
PF0006920=Pkinase=PU(6.9=95.7)
Other Skipping Isoforms:
NA
Associated events
Conservation
Primers PCR
Suggestions for RT-PCR validation
F:
ACGCCGAGCAACTACTACTCT
R:
CCTCACCCAAAGTGCTGACAA
Band lengths:
183-274
Functional annotations
There are 1 annotated functions for this event
PMID: 16371011
Skipping of exon 4 (HsaEX0015698) increases in brain areas affected by Alzheimer's Disease resulting in increased expression levels of clk2tr. Increased expression of clk2 full-length induces skipping of clk2 exon 4. Increased expression of clk2tr does not promote skipping of clk2 exon 4.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]