RnoEX0021838 @ rn6
Exon Skipping
Gene
ENSRNOG00000020500 | Clk2
Description
CDC-like kinase 2 [Source:RGD Symbol;Acc:1359711]
Coordinates
chr2:188480977-188482371:+
Coord C1 exon
chr2:188480977-188481205
Coord A exon
chr2:188481869-188481956
Coord C2 exon
chr2:188482305-188482371
Length
88 bp
Sequences
Splice sites
3' ss Seq
CCCCCCCCACCCCCCGACAGCAG
3' ss Score
6.61
5' ss Seq
GATGTACAA
5' ss Score
-5.41
Exon sequences
Seq C1 exon
CAGCTATGACGATCATTCATCTGACCGGCGGCTATACGATCGACGATACTGTGGCAGCTACAGGCGCAATGACTACAGCCGGGATAGAGGAGAGGCCTACTATGACACAGACTTTCGGCATTCCTATGAATACCATCGAGAGAACAGCAGTTACCGAAGCCAGCGCAGCAGCCGCCGGAAACCCAGACGGCGGAGAAGACGTAGCAGAACATTCAGCCACTCATCTTCA
Seq A exon
CAGCACAGCAGCCGGAGAGCCAAGAGTGTAGAGGACGACGCTGAGGGCCACCTCATCTACCACGTCGGGGACTGGCTACAAGAGCGAT
Seq C2 exon
ATGAAATTGTAAGCACCTTAGGAGAAGGGACCTTTGGCCGAGTTGTACAGTGTGTGGACCATCGCAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSRNOG00000020500_CASSETTE2
Average complexity
S*
Mappability confidence:
100%=100=100%
Protein Impact
ORF disruption upon sequence exclusion
No structure available
Features
Disorder rate (Iupred):
C1=0.578 A=0.367 C2=0.011
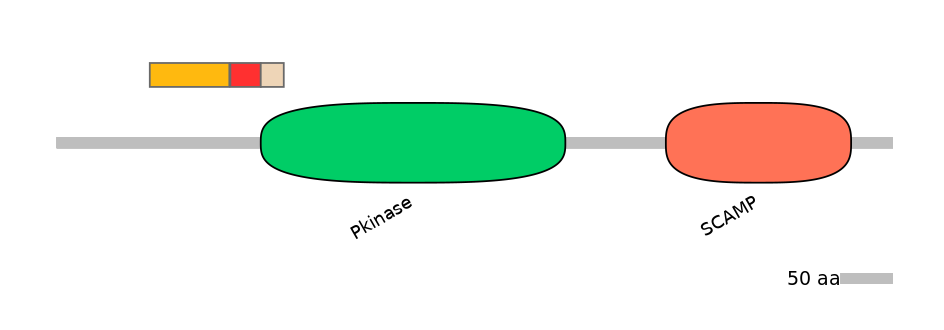
Domain overlap (PFAM):
C1:
NO
A:
PF0006920=Pkinase=PU(0.1=0.0)
C2:
PF0006920=Pkinase=PU(6.9=95.7)
Main Skipping Isoform:
NA
Other Skipping Isoforms:
NA
Associated events
Conservation
Primers PCR
Suggestions for RT-PCR validation
F:
ACACAGACTTTCGGCATTCCT
R:
CACACTGTACAACTCGGCCAA
Band lengths:
179-267
Functional annotations
There are 1 annotated functions for this event
PMID: 16371011
Skipping of exon 4 (HsaEX0015698) increases in brain areas affected by Alzheimer's Disease resulting in increased expression levels of clk2tr. Increased expression of clk2 full-length induces skipping of clk2 exon 4. Increased expression of clk2tr does not promote skipping of clk2 exon 4.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]