GgaEX0008580 @ galGal3
Exon Skipping
Gene
ENSGALG00000006057 | CLK4
Description
NA
Coordinates
chr13:14303756-14305735:+
Coord C1 exon
chr13:14303756-14303981
Coord A exon
chr13:14305175-14305265
Coord C2 exon
chr13:14305669-14305735
Length
91 bp
Sequences
Splice sites
3' ss Seq
GATCTTGGTGCCGTTTCCAGAAG
3' ss Score
7.02
5' ss Seq
GATGTATAG
5' ss Score
-2.49
Exon sequences
Seq C1 exon
TCATTATTTGGACGATAGAACCATAAATGAAAGAGACCATCATGACCGGAGATATGTTGAGGAGTACAGAAATGACTTCTGTGAAGTATATGACCACAGGCATTATCACAGAGACTATGAAAAGAGTCACCATCATCACTATAGCAAATCCTCTGGTCGGAGCAGGAAAAGTAGTCATAAAAGGAAGCATAAGAGGCATCATTGCTCCAGTCACCAATCACATTCG
Seq A exon
AAGAGTCACCGAAGGAAAAGATCCAGGAGTGTAGAGGATGATGAGGAGGGTCACCTGATCTGTGAAAGTGGAGACGTTCTAAGAGCAAGAT
Seq C2 exon
ATGAAATTGTTGCGACTTTAGGAGAAGGCGCTTTTGGAAAAGTGGTGGAGTGCATAGATCATGATAT
VastDB Features
Vast-tools module Information
Secondary ID
ENSGALG00000006057-'7-9,'7-7,8-9
Average complexity
C1
Mappability confidence:
62%=100=25%
Protein Impact
ORF disruption upon sequence exclusion
Show structural model
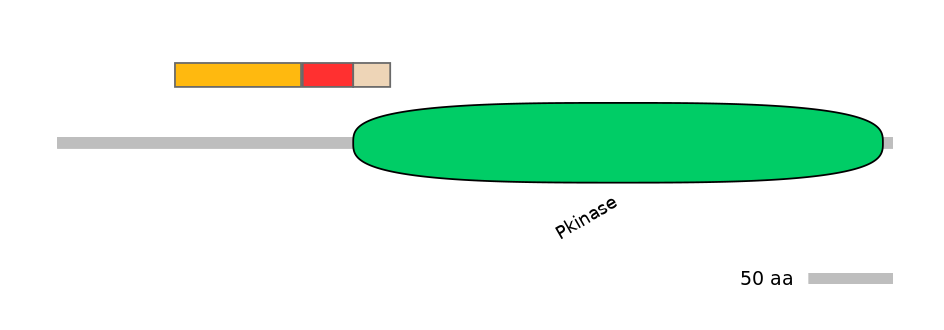
Features
Disorder rate (Iupred):
C1=0.696 A=0.484 C2=0.000
Domain overlap (PFAM):
C1:
NO
A:
PF0006920=Pkinase=PU(0.1=0.0)
C2:
PF0006920=Pkinase=PU(7.0=95.7)
Main Skipping Isoform:
NA
Other Skipping Isoforms:
NA
Associated events
Other assemblies
Conservation
Primers PCR
Suggestions for RT-PCR validation
F:
TGACCACAGGCATTATCACAGA
R:
TTTCCAAAAGCGCCTTCTCCT
Band lengths:
176-267
Functional annotations
There are 1 annotated functions for this event
PMID: 31911676
[CRISPR screen]. Conserved poison exon with negative fitness/survivability impact when depleted: HeLa Depleted at 14 days (FC=0.725, FDR=0.000), PC9 No Change at 14 days (FC=1.106, FDR=0.829), Late Xenograft No change (FC=1.369, FDR=0.260).