MmuEX6079388 @ mm9
Exon Skipping
Gene
ENSMUSG00000001870 | Ltbp1
Description
latent transforming growth factor beta binding protein 1 [Source:MGI Symbol;Acc:MGI:109151]
Coordinates
chr17:75758630-75761978:+
Coord C1 exon
chr17:75758630-75758809
Coord A exon
chr17:75760052-75760138
Coord C2 exon
chr17:75761850-75761978
Length
87 bp
Sequences
Splice sites
3' ss Seq
TTTGTCTCTCCTAAAATCAGCTG
3' ss Score
5.75
5' ss Seq
AAGGTCAGC
5' ss Score
7.09
Exon sequences
Seq C1 exon
ATTCAGGTGTGGATCGTCAGCCTAGAGAAGAGAAGAAGGAGTGCTATTATAACCTCAATGATGCTAGTCTCTGTGATAATGTCCTGGCCCCCAACGTCACCAAACAAGAGTGCTGCTGTACGTCAGGTGCCGGCTGGGGAGACAACTGTGAGATCTTCCCTTGCCCCGTCCAGGGAACTG
Seq A exon
CTGAGTTCACGGAAATGTGCCCTAGAGGGAAAGGTTTGGTCCCGGCTGGAGAATCCTCTTACGACACTGGTGGCGAGAACTACAAAG
Seq C2 exon
ATGCTGATGAATGCTTGCTGTTTGGAGAGGAAATCTGTAAAAATGGTTACTGTTTGAACACTCAACCTGGGTATGAATGCTACTGTAAGCAAGGGACATACTATGATCCTGTCAAATTACAGTGCTTTG
VastDB Features
Vast-tools module Information
Secondary ID
ENSMUSG00000001870-'30-35,'30-34,31-35=AN
Average complexity
A_S
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (Ref)
No structure available
Features
Disorder rate (Iupred):
C1=0.000 A=0.100 C2=0.000
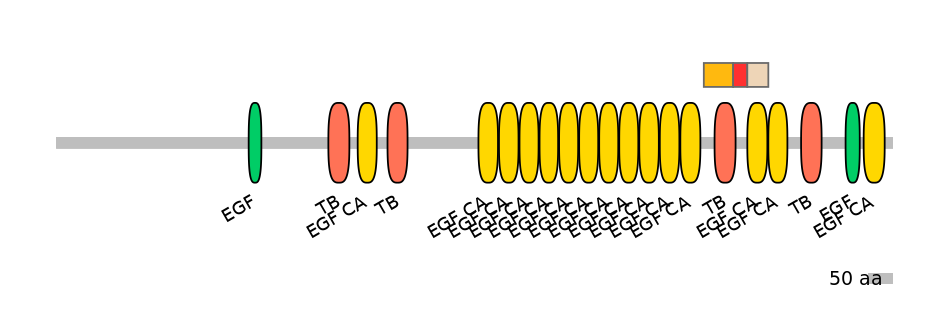
Domain overlap (PFAM):
C1:
PF062476=Plasmod_Pvs28=PD(0.6=1.6),PF0068312=TB=PU(86.4=62.3)
A:
PF0068312=TB=PD(11.4=16.7),PF0764510=EGF_CA=PU(0.1=0.0)
C2:
PF0764510=EGF_CA=WD(100=95.5),PF0764510=EGF_CA=PU(0.1=0.0)
Main Skipping Isoform:
NA
Other Skipping Isoforms:
NA
Associated events
Other assemblies
Conservation
Zebrafish
(danRer10)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
CCCCAACGTCACCAAACAAGA
R:
GCATTCATACCCAGGTTGAGTGT
Band lengths:
172-259
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
Other AS DBs: