MmuEX6082039 @ mm10
Exon Skipping
Gene
ENSMUSG00000042507 | Elmsan1
Description
ELM2 and Myb/SANT-like domain containing 1 [Source:MGI Symbol;Acc:MGI:2685106]
Coordinates
chr12:84158976-84160017:-
Coord C1 exon
chr12:84159875-84160017
Coord A exon
chr12:84159650-84159753
Coord C2 exon
chr12:84158976-84159051
Length
104 bp
Sequences
Splice sites
3' ss Seq
AATGCTTCTGGTGAGTCCAGTGG
3' ss Score
-1.63
5' ss Seq
CTGGTAAGG
5' ss Score
9.68
Exon sequences
Seq C1 exon
ACGAATCAACGTCGGGACCCGGTTCCAGGCAGAAATCCCCATGATGAGAGACCGAGCGCTGGCAGCTTTTGACCCCCATAAGGCTGACTTGGTGTGGCAGCCATGGGAACATCTTGAGAGCAGCTGGGAGAAGCAGAGACAAG
Seq A exon
TGGATGACCTGCTGACAGCTGCCTGTTCAAGCATTTTCCCTGGGGCCGGCACCAACCAGGAGCTGGCCCTGCACTATCTGCATGAGTCCCGGGGGGACATCCTG
Seq C2 exon
GAGGCGCTGAATAAGCTGCTACTGAGGAAGCCCCTGCGGCCTCACAATCACCCACTGGCGACATATCACTACACAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSMUSG00000042507-'21-19,'21-18,22-19
Average complexity
S
Mappability confidence:
100%=100=100%
Protein Impact
ORF disruption upon sequence exclusion
No structure available
Features
Disorder rate (Iupred):
C1=0.061 A=0.000 C2=0.038
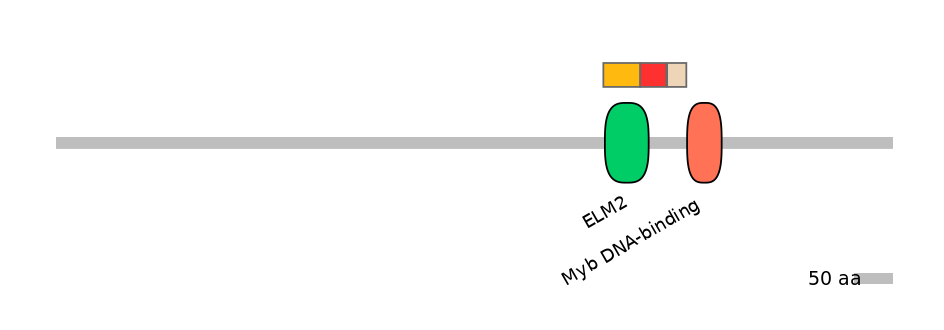
Domain overlap (PFAM):
C1:
PF0144819=ELM2=PU(79.3=93.9)
A:
PF0144819=ELM2=PD(19.0=31.4)
C2:
NO
Main Skipping Isoform:
NA
Other Skipping Isoforms:
NA
Associated events
Other assemblies
Conservation
Chicken
(galGal3)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
CGAATCAACGTCGGGACCC
R:
CTGTGTAGTGATATGTCGCCAGT
Band lengths:
218-322
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Ribosome-engaged transcriptomes of neuronal types
- Neural differentiation time course
- Muscular differentiation time course
- Spermatogenesis cell types
- Reprogramming of fibroblasts to iPSCs
- Hematopoietic precursors and cell types