RnoEX0009390 @ rn6
Exon Skipping
Gene
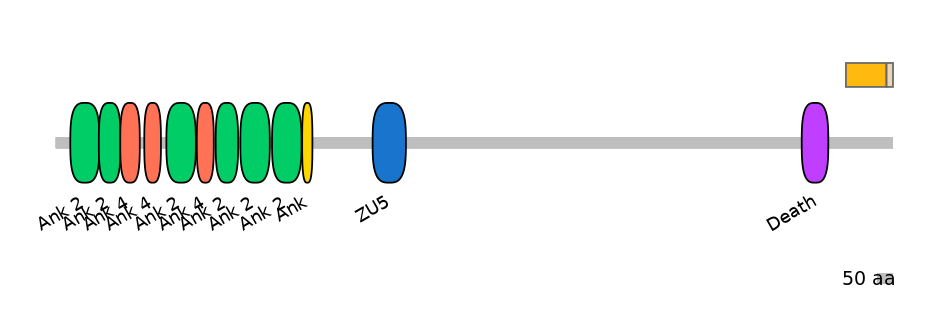
ENSRNOG00000053288 | Ank3
Description
ankyrin 3 [Source:RGD Symbol;Acc:620157]
Coordinates
chr20:20454767-20471282:+
Coord C1 exon
chr20:20454767-20455145
Coord A exon
chr20:20469867-20469923
Coord C2 exon
chr20:20471197-20471282
Length
57 bp
Sequences
Splice sites
3' ss Seq
CTCTGTGTTGCTTCCAACAGATC
3' ss Score
10.92
5' ss Seq
GTGGTAATG
5' ss Score
4.46
Exon sequences
Seq C1 exon
CTCTGACCAGGTTCGGGATCCCATTACCTCATACCTCACAGGAGAAGCTGGGAAGTTCGAAGCAAACGGAAACCACGCAGAAGTCATTCCAGAAGCAAAGGCCAAGGCCTACTTTCCTGAATCCCAAAACGATATAGGGAAACAGAGCATCAAGGAGAACTTGAAACCAAAAACACACGGATGTGGTCGCGCTGAGGAACCAGTGTCGCCCCTCACAGCCTACCAGAAATCTCTAGAAGAAACCAGCAAGCTTGTCATAGAAGACGCACCTAAACCCTGTGTGCCTGTCGGCATGAAAAAGATGACCAGGACTCCGGCTGACGGCAAAGCCAGGCTCAACCTCCAGGAAGAGGAGGGGTCCGCCAGGTCAGAGCCTAAG
Seq A exon
ATCACTCGGAAGGTAATAAGACGGATTGGTCCACAAGAGAGGAAGCAGGATGATGTG
Seq C2 exon
CAGGGAGAAGGCTATAAGGTGAAAACAAAGAAGGAAATCCGGAATGTGGAGAAGAAAGCCCACTAGTGACGGTGACGTCAGTCCAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSRNOG00000053288_MULTIEX1-4/4=C1-C2
Average complexity
ME(1-4[99=100])
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (No Ref)
No structure available
Features
Disorder rate (Iupred):
C1=0.930 A=1.000 C2=1.000
Domain overlap (PFAM):
C1:
NO
A:
NO
C2:
NO

Associated events
Conservation
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
AAAGATGACCAGGACTCCGGC
R:
GCTTTCTTCTCCACATTCCGGA
Band lengths:
141-198
Functional annotations
There are 1 annotated functions for this event
PMID: 21223964
Ankyrins-G coimmunoprecipitated with plectin and filamin C from skeletal muscle tissue extracts. Muscle-specific OTBD domain (coordinated inclusion of MmuEX1003810, MmuEX1003809, MmuEX0004908) is required for ankyrin-G binding to plectin and filamin C. Isoforms are differentially expressed during myogenesis.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]