RnoEX0018971 @ rn6
Exon Skipping
Gene
ENSRNOG00000002841 | Cdc42bpa
Description
CDC42 binding protein kinase alpha [Source:RGD Symbol;Acc:621406]
Coordinates
chr13:98415631-98419232:+
Coord C1 exon
chr13:98415631-98415735
Coord A exon
chr13:98417413-98417520
Coord C2 exon
chr13:98419149-98419232
Length
108 bp
Sequences
Splice sites
3' ss Seq
TGTCTGTTTCACCCCAGCAGCAC
3' ss Score
5.9
5' ss Seq
CTGGTACTT
5' ss Score
4.74
Exon sequences
Seq C1 exon
ACTGAGCCCGTTGACAACACTTACATACGGAATCCGAGCGTCAAGCTTTACATCCAGCAAAGGCCCACATCTCCTTCCACACCTAGTGAAGCTGAGCCAGTTAAG
Seq A exon
CACTGTTTCTTCTCTGCATGCTCGGGTCTGTCATGGCCTCCCGGTCCTGTGCTTACTCTTGCACCGAGCCTACGTTGTCTTCGTCACTGCAGCCCTGAAACAGTTCTG
Seq C2 exon
ATTGCAGACTCTGCTCCGCTTCCAGTTCACACACCAACTTTAAGGAAAAAGGGATGCCCAGCTTCAGCTGGTTTTCCACCTAAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSRNOG00000002841-'45-45,'45-44,46-45
Average complexity
C3
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (No Ref)
No structure available
Features
Disorder rate (Iupred):
C1=NA A=0.000 C2=0.443
Domain overlap (PFAM):
C1:
NA
A:
NO
C2:
NO
Main Inclusion Isoform:
ENSRNOT00000003837fB2725
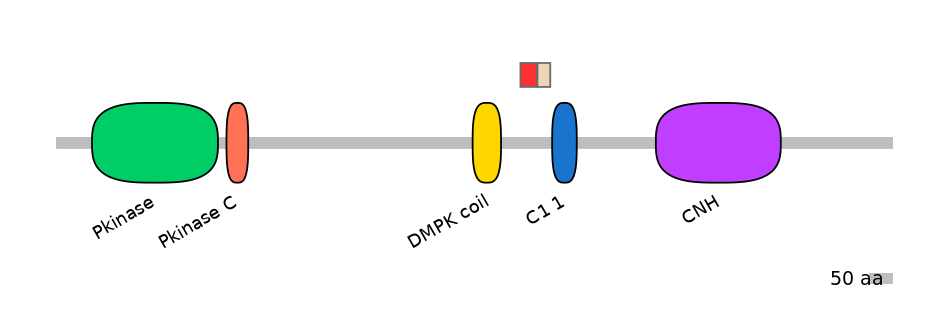
Main Skipping Isoform:
ENSRNOT00000003837fB2728
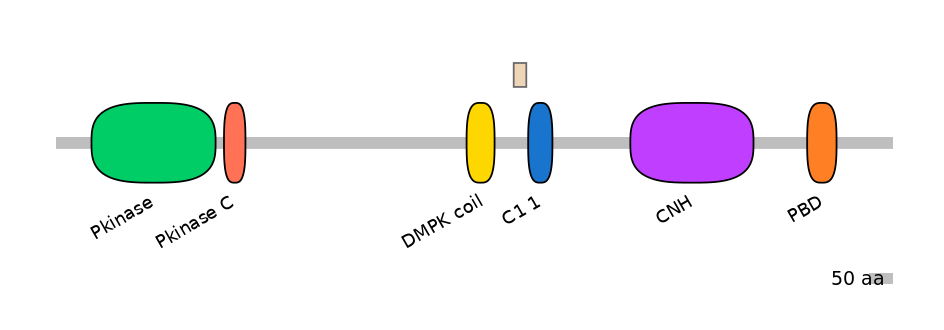
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Conservation
Human
(hg19)
No conservation detected
Mouse
(mm9)
No conservation detected
Cow
(bosTau6)
No conservation detected
Chicken
(galGal4)
No conservation detected
Chicken
(galGal3)
No conservation detected
Zebrafish
(danRer10)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
CTGAGCCCGTTGACAACACTT
R:
GTGGAAAACCAGCTGAAGC
Band lengths:
183-291
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]