RnoEX0078584 @ rn6
Exon Skipping
Gene
ENSRNOG00000015049 | Scn5a
Description
sodium voltage-gated channel alpha subunit 5 [Source:RGD Symbol;Acc:3637]
Coordinates
chr8:128198493-128203242:-
Coord C1 exon
chr8:128202802-128203242
Coord A exon
chr8:128201361-128201519
Coord C2 exon
chr8:128198493-128198613
Length
159 bp
Sequences
Splice sites
3' ss Seq
TCATGGGGTCTTTTCAGCAGGAA
3' ss Score
5.89
5' ss Seq
GAGGTAACG
5' ss Score
9.65
Exon sequences
Seq C1 exon
GTCCTGAATCTCTTCTTGGCCTTGCTGCTCAGCTCCTTCAGCGCAGACAACCTCACAGCCCCTGACGAGGATGGGGAGATGAACAACCTCCAGCTGGCCCTGGCTCGCATCCAGAGGGGCCTGCGCTTTGTCAAGCGGACCACCTGGGACTTCTGCTGCGGGATCCTGCGGCGGCGACCTAAGAAGCCCGCGGCTCTTGCCACCCACAGCCAGCTGCCCAGCTGTATCACCGCCCCCAGGTCCCCACCACCCCCAGAGGTGGAGAAGGTGCCCCCAGCCCGCAAGGAAACACGATTCGAGGAGGACAAGCGACCCGGCCAGGGCACCCCTGGGGATTCGGAGCCTGTGTGTGTGCCCATCGCCGTGGCTGAGTCAGACACTGAAGACCAGGAAGAGGATGAAGAGAACAGCCTTGGCACAGAGGAAGAGTCCAGCAAACAG
Seq A exon
GAATCCCAAGTTGTGTCTGGTGGCCACGAGCCCTACCAGGAGCCCAGGGCCTGGAGCCAGGTGTCAGAGACCACGTCCTCTGAAGCTGGGGCCAGTACATCTCAGGCAGACTGGCAGCAAGAGCAGAAAACGGAGCCCCAGGCCCCGGGGTGCGGTGAG
Seq C2 exon
ACCCCTGAGGACAGTTACTCCGAGGGCAGCACAGCTGACATGACCAACACCGCCGACCTCCTGGAGCAAATCCCAGACCTTGGTGAGGACGTCAAGGACCCAGAGGACTGCTTTACTGAAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSRNOG00000015049_CASSETTE1
Average complexity
S*
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (Ref)
No structure available
Features
Disorder rate (Iupred):
C1=0.531 A=1.000 C2=0.683
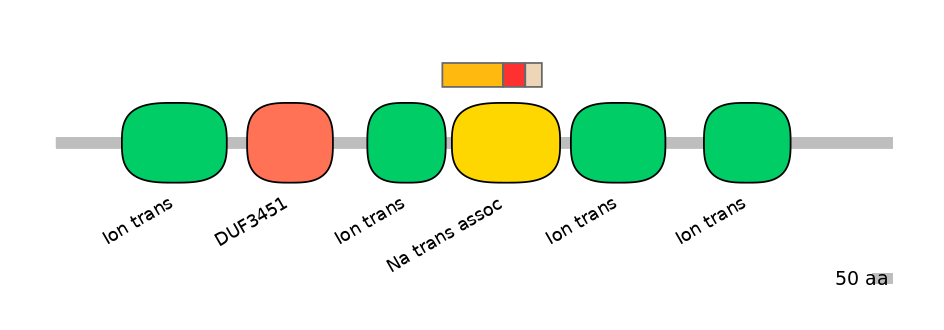
Domain overlap (PFAM):
C1:
PF0052026=Ion_trans=PD(4.2=5.4),PF065128=Na_trans_assoc=PU(46.9=83.7)
A:
PF065128=Na_trans_assoc=FE(19.8=100)
C2:
PF065128=Na_trans_assoc=FE(15.3=100)
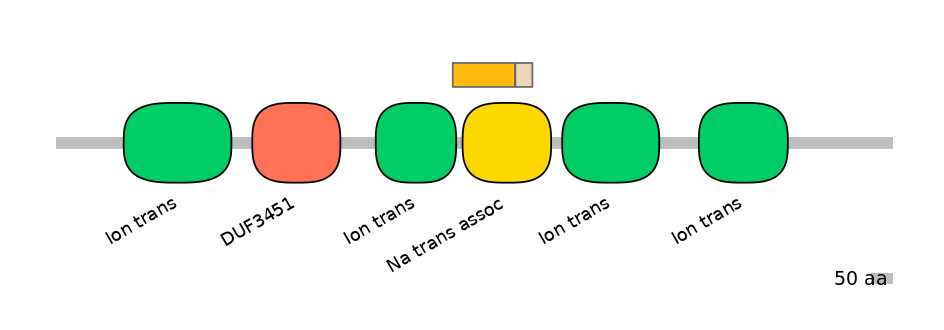
Other Skipping Isoforms:
NA
Associated events
Conservation
Chicken
(galGal4)
No conservation detected
Chicken
(galGal3)
No conservation detected
Zebrafish
(danRer10)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
CACGATTCGAGGAGGACAAGC
R:
TTGACGTCCTCACCAAGGTCT
Band lengths:
247-406
Functional annotations
There are 1 annotated functions for this event
PMID: 21552533
Negative results. Nine Nav1.5 (SCN5A) splice variants are known. Four of them, namely Nav1.5a, Nav1.5c, Nav1.5d, and Nav1.5e, generate functional channels in heterologous expression systems. In the present study, the authors systematically investigated electrophysiological properties of mutant T1620K channels in the background of all known functional Nav1.5 splice variants in HEK293 cells. This mutation is associated with two cardiac excitation disorders: long QT syndrome type 3 (LQT3) and isolated cardiac conduction disease (CCD). When investigating the effect of the T1620K mutation, the authors noticed similar channel defects in the background of hNav1.5, hNav1.5a, and hNav1.5c. In contrast, the hNav1.5d background produced differential effects: In the mutant channel, some gain-of-function features did not emerge, whereas loss-of-function became more pronounced. In case of hNav1.5e, the neonatal variant of hNav1.5, both the splice variant itself as well as the corresponding mutant channel showed electrophysiological properties that were distinct from the wild-type and mutant reference channels, hNav1.5 and T1620K, respectively. In conclusion, these data show that alternative splicing is a mechanism capable of generating a variety of functionally distinct wild-type and mutant hNav1.5 channels. Thus, the cellular splicing machinery is a potential player affecting genotype-phenotype correlations in SCN5A channelopathies. (About variants: Variant a and c differs from the canonical variant by skipping exon 18 (HsaEX0056556), and including Q1077 (HsaALTA0007521-2/2), respectively. Negative results for those. Variant D seems to use alternative splice sites in exon 17 (not in VastDB), and variant E uses the neonatal version of the ME exon (HsaEX0056558), while the other variants use the adult version (HsaEX0056559)).
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]