RnoINT0168111 @ rn6
Intron Retention
Gene
ENSRNOG00000017249 | Zfp366
Description
zinc finger protein 366 [Source:RGD Symbol;Acc:2322904]
Coordinates
chr2:29494170-29498664:+
Coord C1 exon
chr2:29494170-29494344
Coord A exon
chr2:29494345-29497062
Coord C2 exon
chr2:29497063-29498664
Length
2718 bp
Sequences
Splice sites
5' ss Seq
AAGGTACTC
5' ss Score
7.46
3' ss Seq
ATTCTGCTCTGCTCCCGTAGGTC
3' ss Score
10.41
Exon sequences
Seq C1 exon
CTGTGCGGGAAGGAATTCAACAGGATGCACAACCTGATGGGCCACCTGCACCTGCACTCGGACAGCAAGCCCTTCAAGTGTCTCTACTGTCCCAGTAAGTTCACGCTGAAGGGGAACCTGACCCGCCATATGAAGGTCAAGCATGGCGTGATGGAGCGAGGCCTTCACTCTCAAG
Seq A exon
GTACTCACTCCCTCCACTGTGGCTGGAAGCCCTGTCCACCACACCCGGGGGGGTGCACAGGGAAGAGCAGTGTGCTTTTATGGGGGCACTTCTTGGGGGGGTGGGATTATCCTTGCCCAGAATGGGGGTGTATTTTAATGACCACAGCAGAGCAGCAGAGCAGGCAAGAGTGAAAGGGTCACAGAGCAACTCTGGAGCACAGTTATGATGGCAGTGGTCCCGTCTCCTTTGTCTCAGGCCATGCGTGGTATGCTCCATCTCTTTAAATCTTCAGTCACCCCCAGCTAGAGTGCCTCTGTTGTCCCCATTTCGCAGGTTGGAAATCTGAGACTCCCAAAGTTATTCTATCTTGATTGCATTTCAACTTTATTATATATAGTCATTATTATATTATTACCCGTACTTTTCACACAGATAGACAACTCGGGGTTTTTCTTCCATTTTCAAAACAAATGATGGGTCTGCATCCCATCCGGCCGCAGAGATGTTTGCTACCCTTTCTGTCCACACAAGGGCAGCTTTCCCATTGCCCCAGCTCCTACATGAGCAGGGGTTTGTGTCTCTTGACTGAACGAATGAAAAGCTCTTGTTTATTTAATCTCCTTAGCTCCAGACCCTTGTTGGGGAAGACTTTATGTTCCAGGTCCCTTGAGTGCAGCACGCTACTTAGTCACCTCACCAGCCAGCCCTACAAAAACAAAACAAACAAACAAAGCCAGACAATAAGTGCAACCCCCACCACCACCACACACACATATACCAAGCGGATGCAGGGACAAAGTATGGAGAATGAGAGAGGGAGCACCTGGGCCCTCTAGGTCTGTGTGAAGATCGAAGACAGTCACACATAGTGTTCATAAAATTACATCAACAGTTTTTGACCAGAAAAACGGTGACAACTGAGGGTAAGACTTAGCAGCAGCCTCCTCAGTCACCTCTTCAAGTCCGTGAGAGAGCCTGTCCTCATCTTGTGTCTGAGGAGAACTTGCAGGTGCCACATAAACTTCCTGTGGCCGGCAGAAGTTTGCAGGCAGAGCTGGGACTGATGTCAAGCCTCCATTCGCTCACCATCCCAGGCAAAACCATGGCTGACTTGTTTGTCTTTACTGCTTATGAAGTAAGGTGCAAACACTGATAAAAACCCATGTCTGAGCAAACAACAAAAACCAAAATAAGCATAGACTGGGGAGATTTTACAACGGCTCTGAGTTTATGGTGACCCCGGGGCTCACAGGCAACTATGGTCACTACAAGTGAGTTTATGCAAAAACCTTTTAAAAGTACTGGTTAAAAAGAGAATGTTTCTCTTTTCCTTGGGATGCCCTTTCTCTTTACTCATTTATTGACTCTGGTCCGCCAAGCCCCTTCAAGGCAAGACATGGATTTTTCTAAAAGTAGAGTTTGTTTCATCACTTGGGTGGGAAACAGTTATAGCAAGAAGAAGAAGCCATCCACTAGAAGCCTTATGTGGTCATTGTGAATTACCCCGCAGGGAAACTCATATCCGAATAGACATATTGTGGCTCTGTCACTGTGGGGCAAGGAGAGCATCACTACGTAACAGAGGATTGCTTTGAAAAACAATTACTGGTTTTTCAACAATTCTGGTGGCTTTATCTATACATGGTAATTTTTCTTGTGGCTCAAAAAAAAAAATCATTAGAAAGATGAAAAGATTCCTCTTGGCCCCTGACAAGATTAAAGCTATGGACAGCAGGCATCATTGTGCCTCAAAGCCCCGACCCTTCCTTTCTTTCCTCCCACTCCAGCCTGCCACCTGCTGTATCCTGGACACCTCCCTTCCTGTTTCTCAAACCCTCAGTGTCCCTGCTGTGTCTGACGGTCTGGTTTGGGTCATGGAAGGCAGAGGCCTTGAAATGAGCTGCAGTCTGTGCCCAGTTCTAAACCCTAAGCCTGTGCTTCTAGCAAAAAGTGAAAGGGACCCTCCCCCATGTCACCCTGTCCATTGCAGGGACATTGGATCCAGATCGGGGACTTGGGAGCTCCTCAGAGAACCTCTACTTTGTGCATAGGGACCACAGGCTGTGGAGTTGCCTAAGTGTGTTTGTCTTTGCTGTGGCTTTATTTGTTTGAGGTCTTGAGACCTCAATATTTATTAAGGTCTACATGTTCTCTGTGAAAAGAAAATGAGAAATACTTGAGAAGTGAAGAGAAAGTCCAGTAGCCACTTCAGAGTTTTGAGATCTGATTCTTTATCCTTCCACACAGTTCTGATAAGGAAAAAGGGGAAGAGACATAAGTGGGGAAGGGGAGGGAGGGAGGGAGAGATTTTAATTTAGAATACACAGAGAATATGTGTATGGATTCTCATGTATGTAGGCATTTTTAATCAGCCAGACACAGTTCATTGTGTAGTTCCATTTATGTGTTTAAACCAAAATTTAGATACTATTTTTTCACATGTAGTATAATAAAAAAAAAGTGTTTCGCACATACTCCGTGTCCCTCCTCCCTTACTTTTTCATATTTTAAATTCTCAGTAGAGAGACGCTAAGTTGGGGCACAAGGTGCCAGGTTTGCCTTACAGAGTGCTATGGATCACAAGCTCCCCACGCCAGAGTCCTTAATGGGAACTATTCCCCCCTCCCCCCTCCCCGGCCCCCAACCCCATCCCTGTTTTTAAAATGAACAAGCAAAATGCCTAAGGGGCCTCCGGCATAACTTTTCTCAGACATAAATTCTGCTCTGCTCCCGTAG
Seq C2 exon
GTCTGGGTAGGGGACGACTCGTCCTGGCACAGTCCACTGGTGCGCTGAGGAATCTGGAGCAGGAGGAGCCGTTTGACCTCTCCCAAAAGCGCAGTGCAAAGGGGCCAATGTTCCAGTCCGACATGGACAGCACGCAGGGCTGCCTCTACCCAGAGGAGGAGGAGGAAGAGGAGGAGGAGGCAGATGAAGAGGACAAGTGCTATGACGTGGAGCCCTATAGCCCCACCCTGGCCTTGGAGAGCCAGCAGCTCTGTGCGCCTGAGGATCTGTCCACCAAGCAAGAGCAGGCCCCGAAAGGCCCTGGAGAAGGATGCCAGGATCAGGACACTCCAGAAGAGCCGCAGGAGGACAGTAGCGAGGACCATGAGAGTAGAGACATAGACTGTGAAGTCAGAGACGAGCGACTGAGCAGCAGGCTGTTGCAAAGCGGCGGGCAGGGCCCCTCGTTTTCTGATTACTTATACTTCAAGCACAGAGATGAGGGTTTAAAAGAATTACTGGAGAGGAAAATGGAAAAGCAAGCAGTCCTTTTGGGTATCTAAGTAAAACGGCTTTAAAGTTACATTTGGAAAAGGAGATCGAGGCAGTTCAACTATAGCTTTTCGTGCAAACTGTCATTTGCTGCAAAGGGGAGAGAGGCGCCAATCTTCACCAATGGCTCAGACTGAATGCGTGGGTAACACGCACACTTCTCAGGTCCTTCCTTGGGCACTAGACCCTTTGGTGTTATTTTACACATCACCCGCATGATGAACTGTGCCTCCTTCTAGATACCTAACTGCAAGCTGGACCTCGAGGTGCTTTTAGACACTGTTTTCTCCTAACATTCAGTGGCGCAAACATCTTTCTAGTTGCAAGACTGCGGTGCTCGAGTTATCTTACTTAGTAACTAGTTAGCCGCATTTATGAAGGCTACAGTATCATAGATGAAAATATTAGTAGTTGTATATATAAAAATATAGCCTCTATGAAATAACAGAAACACAGGAAATGATGCAAGAGACCAACTGTTGGGAAGGTTCGAGTGGTTCCTGCAGACGTGAGGTTGGGTTTCAGGAGAAGGAAAATCGTTTTTTCTAAGACTGAACCAGATGATGCTTAAATATCTGGTTGAGTCCTCCCCTACTTCAGCTTTTCTGGTCAGTGGGACATTTGAGACACCCCTGGAGACATCTGGATAAGCTTGGGGGTCCCAGTTCTTCAGATCAAACCCCTCACAAGAAGAGCTCCTTCCCCTTGCTCTGAGGCCCCATGCCCCATGAAGCAACTGAAGGGAGAGTTGCCTTTCAGAGTGAGCAGTGGAGCTTGGGCTATGGAATGGGGTTTAGGACTGCTCCCTAAAACAAACTTTTATCTGGGTAGAGGAATGATCACCCGCCCCTCGCTATAAGCAAGACCTGAGGTATCCTCTGAGAGCTGGCTACATGTAGGAATCATTCAGTAGATCCTTCAGAAGGCACTGATGTAGCTGGGCATGGTATCTCACAACTGTAATCCCAGCACTTGGGAGGCTGAGGGGGATTATGAGGTCAAACCTTCTCTCAAAGAAAAAAGGTGGAAGTAAGCGAAACCCTGTAAGGTTCTTGGTGAATCCACCTGGAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSRNOG00000017249:ENSRNOT00000023180:4
Average complexity
IR
Mappability confidence:
NA
Protein Impact
Alternative protein isoforms
No structure available
Features
Disorder rate (Iupred):
C1=0.119 A=NA C2=0.778
Domain overlap (PFAM):
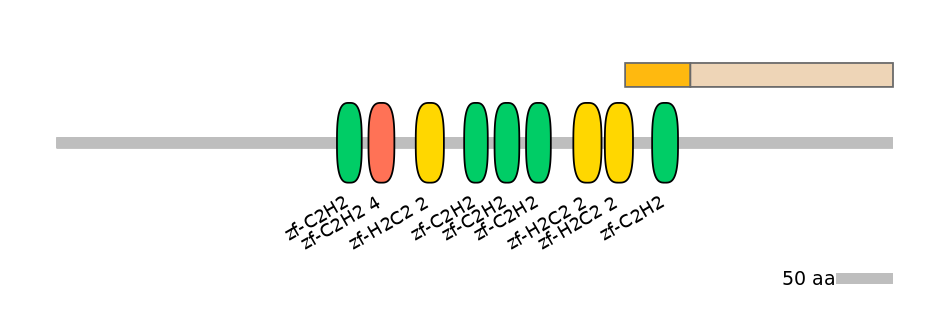
C1:
PF134651=zf-H2C2_2=PD(26.9=11.9),PF0009621=zf-C2H2=WD(100=40.7)
A:
NA
C2:
NO
Main Inclusion Isoform:
NA
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Conservation
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
No suggested primer sequences
R:
No suggested primer sequences
Band lengths:
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]