BtaEX0010330 @ bosTau6
Exon Skipping
Gene
ENSBTAG00000002411 | CTNND1
Description
catenin (cadherin-associated protein), delta 1 [Source:HGNC Symbol;Acc:HGNC:2515]
Coordinates
chr15:82318658-82319680:+
Coord C1 exon
chr15:82318658-82318811
Coord A exon
chr15:82319170-82319187
Coord C2 exon
chr15:82319612-82319680
Length
18 bp
Sequences
Splice sites
3' ss Seq
CTTGGGTGACGCGCTGGAAGATG
3' ss Score
-1.64
5' ss Seq
GAGGTGAGT
5' ss Score
10.03
Exon sequences
Seq C1 exon
CTTGTCGAGAACTGCGTCTGCCTCCTTCGGAACCTGTCCTATCAAGTTCACCGGGAGATCCCGCAAGCAGAGCGCTACCAGGAGGCGCCTCCCAGCGTCGCCAACAGTACCGGGCCGCATGCTGCCAGCTGCTTTGGGGCCAAGAAAGGCAAAG
Seq A exon
ATGAGTGGTTCTCCAGAG
Seq C2 exon
GGAAAAAACCCACAGAGGATCCAACAAACGATACAGTGGACTTCCCTAAAAGAACTAGTCCTGCTCGAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSBTAG00000002411_CASSETTE1
Average complexity
S
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (No Ref)
No structure available
Features
Disorder rate (Iupred):
C1=0.365 A=0.571 C2=0.708
Domain overlap (PFAM):
C1:
NO
A:
NO
C2:
PF0051418=Arm=PU(2.5=4.2)
Main Inclusion Isoform:
ENSBTAT00000043778fB1958
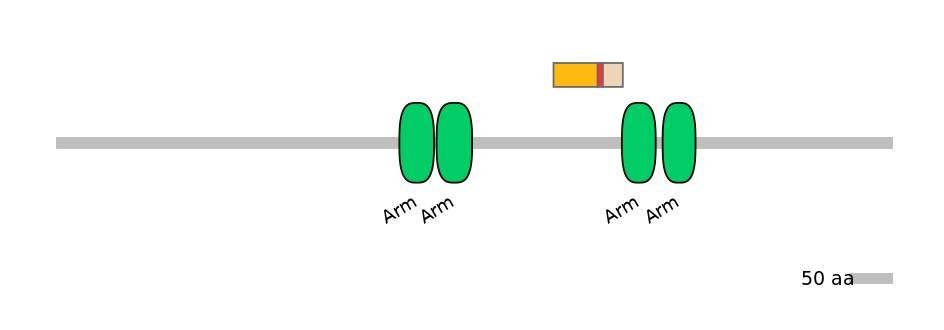
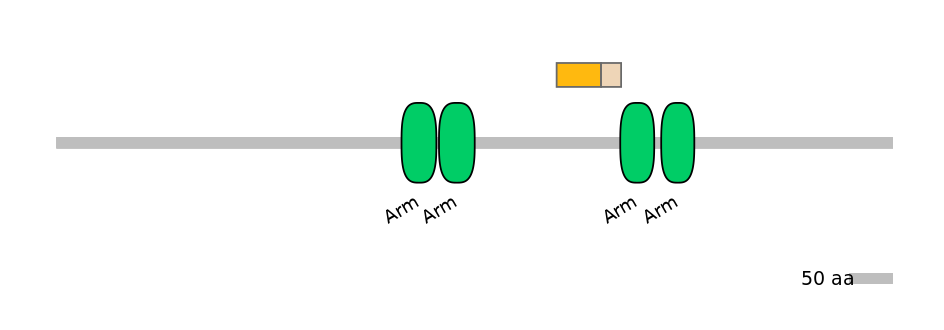
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Conservation
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
AGAGCGCTACCAGGAGGC
R:
TCGTTTGTTGGATCCTCTGTGG
Band lengths:
117-135
Functional annotations
There are 1 annotated functions for this event
PMID: 32423416
The authors hypothesized that AltTEM regulation was responsible for a protein localization change in the Ctnnd1 protein (aka p120). Validation experiments using Western blot analysis of p120 in nuclear/cytoplasmic fractions for both cell types confirmed the increase of nuclear levels of the protein in OPCs in comparison to NPCs, where a cytoplasmic retention of p120 was observed. Higher inclusion, higher nuclear localization.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Pre-implantation embryo development