DreEX0023092 @ danRer10
Exon Skipping
Gene
ENSDARG00000012405 | col1a1a
Description
collagen, type I, alpha 1a [Source:ZFIN;Acc:ZDB-GENE-030131-9102]
Coordinates
chr3:23095661-23104136:+
Coord C1 exon
chr3:23095661-23095714
Coord A exon
chr3:23095800-23095844
Coord C2 exon
chr3:23104029-23104136
Length
45 bp
Sequences
Splice sites
3' ss Seq
CATGCGGCTTTGTTTATTAGGGT
3' ss Score
7.81
5' ss Seq
ATGGTAAGT
5' ss Score
11.01
Exon sequences
Seq C1 exon
GGATTCAGCGGTCTAGATGGAGCTAAGGGGGATGCTGGCCCTGCTGGACCTAAG
Seq A exon
GGTGAGCCTGGTGCACCTGGTGAGAATGGAACTCCTGGTGCCATG
Seq C2 exon
GGACCCGCTGGAGCTCGTGGAGACAAGGGTGAGACTGGTGAGGCTGGTGAGAGAGGCATGAAGGGACACAGAGGATTCACTGGAATGCCAGGACCCCCTGGTCCTCCT
VastDB Features
Vast-tools module Information
Secondary ID
ENSDARG00000012405_MULTIEX1-6/40=5-38
Average complexity
C3
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (Ref)
No structure available
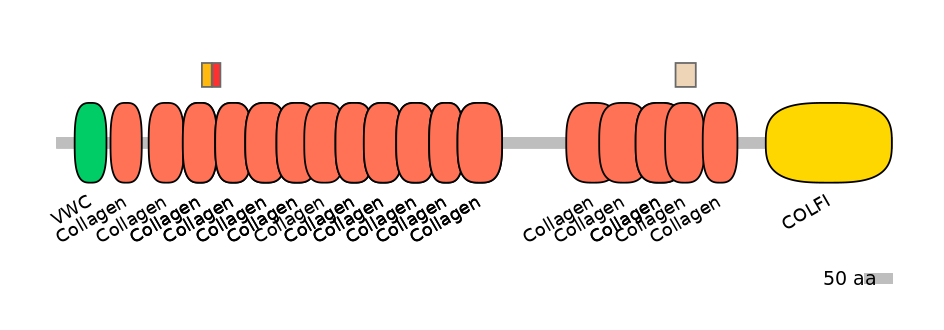
Features
Disorder rate (Iupred):
C1=1.000 A=1.000 C2=1.000
Domain overlap (PFAM):
C1:
PF0139113=Collagen=FE(28.3=100)
A:
PF0139113=Collagen=PD(13.3=53.3),PF0139113=Collagen=PU(14.1=60.0)
C2:
PF0139113=Collagen=PD(17.6=41.7),PF0139113=Collagen=FE(50.7=100)
Main Skipping Isoform:
NA
Other Skipping Isoforms:
NA
Associated events
- DreEX0023059
- DreEX0023060
- DreEX0023061
- DreEX0023062
- DreEX0023063
- DreEX0023064
- DreEX0023065
- DreEX0023066
- DreEX0023067
- DreEX0023068
- DreEX0023070
- DreEX0023071
- DreEX0023072
- DreEX0023073
- DreEX0023074
- DreEX0023075
- DreEX0023076
- DreEX0023077
- DreEX0023078
- DreEX0023079
- DreEX0023080
- DreEX0023081
- DreEX0023084
- DreEX0023085
- DreEX0023086
- DreEX0023087
- DreEX0023091
- DreEX0023093
- DreEX0023094
- DreEX0023095
- DreEX0070576
- DreEX0070577
Conservation
Chicken
(galGal3)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
TTCAGCGGTCTAGATGGAGCT
R:
CCAGTGAATCCTCTGTGTCCC
Band lengths:
134-179
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]