DreINT0149988 @ danRer10
Intron Retention
Gene
ENSDARG00000008568 | slc25a24
Description
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 [Source:ZFIN;Acc:ZDB-GENE-040912-183]
Coordinates
chr20:34751037-34753130:-
Coord C1 exon
chr20:34752980-34753130
Coord A exon
chr20:34751818-34752979
Coord C2 exon
chr20:34751037-34751817
Length
1162 bp
Sequences
Splice sites
5' ss Seq
TGGGTAAAC
5' ss Score
3.32
3' ss Seq
TTTTCTTTTATTCTTCTCAGCGT
3' ss Score
9.26
Exon sequences
Seq C1 exon
ACCTTGAAGAACACCTGGTTGTCTCACTATGCTAAAGACACTGCTAACCCAGGAGTGCTGGTTCTCCTCGGCTGTGGGACCATTTCCAGCACATGCGGACAGTTGGCCAGTTACCCTCTCGCTCTGATCCGCACACGCATGCAGGCCATGG
Seq A exon
GTAAACACATTAACAATCTACACAGTACACACTGCTGTTTTAAAAAAAAAGAATATTCTGCTCATCCATGCTGTGTTTATTTGATCAGAAATACAGTAAGAGGTACAAAAATAGGTACAATGGTACAATTATTACAGACATATTATTACAAGTTAATATTTTCTTAGGTAATGTATTTTCAAATCATTTTTAAAATAATTATTCTAGTCTTCAGTGTCACACGAGCCATCAGAAATCATTCAAGTACCATCTTCTCAAGAAATCTTTGAGATTCTGATTCTGACAACTATTTGGTGGCTGCATATTATTGTGTAAACTGTGATACAGTCTTGTTCAAAAACTATTCTCTAATATTTATCAAGGACATATTACATTTTTCAAGAGTGAAAGTAAAGAGATTTTAATTTAAAAACACAACAACAGGTTATGTTTTAAGTAGATTCTGTTATTTTGTATTCTGATATTATTTTCATCAAAAAATAGTAAGTAAAATTGTAAAAAGTGATATAAAAAGATGTAAGCAGGAATTATAAGTACTATATAATAATTAATGAATGAGTGCTAATGATCATTTTAACATTCATTCATTCAATTCCCTTCGGCTTACTGGCTCATTTATCAGAGGTCACCACAGCGGAATGAACCGCCAACTATTCCGACATATGTTTTATGCAGCAGATGCCCTTCCAGCTGCAGCCAAGTGCTGGGAAACACTCATACACTCATTCACACACATACACTACAGCCAATTTAGTTTATTCAATTCAGCTTTACCGCATGTCTTTGATCCAGAGGAAACCCATGCCAACACAGGGAGAACATGCAAGCTCCACACAAAAATGCCATCTGACCCAGTGTGAACTCGAACCAGTGGCCACTGAGCCACCGTGCCGCCCATTATTGAAACAATAGAATATTGGAAACTCGAGTAATGATGCTGAAAATTCAGATTAAATCGCAAAAGAATAATTTACAGTTTATATTAAATTCAATAATAAAACAATTACTTTAAATGTAAAGTTTTTTTCAATGTTTTTGAGCCTTGATGAGCACAATATGCTTATTTTAAAAACCTTACAAATTCCTACTGATCCTGAATTTTTTGGTGATCAACACTTTTTGCGAATGTGATTTCCTGATGTTTTTCTTTTATTCTTCTCAG
Seq C2 exon
CGTCAATGGAAGGATCAGAGCAGGTGTCTATGTCCAAATTAGTGAAGAAGATCATGCAGAAGGAGGGCTTTTTTGGTCTCTATCGTGGGATCCTGCCCAACTTCATGAAAGTGATCCCCGCTGTCAGCATTAGTTATGTGGTGTATGAGTATATGAGGTCTGGGTTAGGCATCTCTAAATGATGAATGTCTTGCATGTTTTTACACGCACAAATTTCATTCACTTCTCTTTGAAAACACCCTCCATGATGGGACAAAATGTTTTGTTCACTTACCAAAGTAAAAAAAAAAGAAGCCACGATGGTGTCTTGCATTGTGGTCAGGGCCCAACCTAATTCTTGTTTACACTGATGTATAACTGCACCTTTATATGTAATCGATACATTTATGACATGTTTTACAATTTGTATGATTTTGTCTTGATATTCATGTAATATCTGAGCAAACGATGAAGCCATTGTGCACTATATAAATGTGTGTATATATATAAATGAATATAAATGTTTAGGTGAATTAGGATGGGCCACAGTCGAGTCTTTACACTATGCAGAAAAATATATATATCCACATTTAAAAAATGAGAAAAACAGAATCAATGCAAATCGGGGAGCCACGCAATGACAAGAGCTTAGTGTGCCTTATATCATTAGTTCAGTCTTTGTGAGGGTCTCACTAATTCCTGACCTGGAAAGATCAATGTTATTGAACTTGTGTTGCCTTTTGATCCCCATTGTTTTGTTATTTTAGAAACTCCTGTTTACATTAAAGGTTGTATTTTAATA
VastDB Features
Vast-tools module Information
Secondary ID
ENSDARG00000008568:ENSDART00000033325:9
Average complexity
IR
Mappability confidence:
NA
Protein Impact
Alternative protein isoforms
No structure available
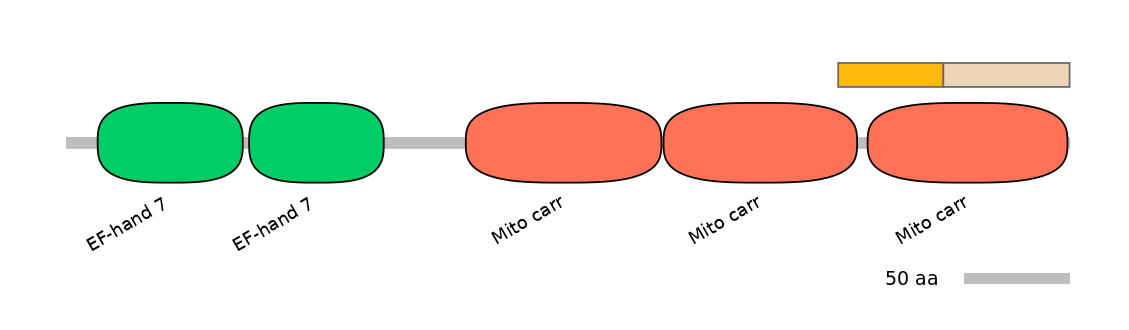
Features
Disorder rate (Iupred):
C1=0.000 A=NA C2=0.000
Domain overlap (PFAM):
C1:
PF0015322=Mito_carr=PD(9.7=17.6),PF0015322=Mito_carr=PU(37.5=70.6)
A:
NA
C2:
PF0015322=Mito_carr=PD(61.5=96.7)
Main Inclusion Isoform:
NA
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Conservation
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
TTACCCTCTCGCTCTGATCCG
R:
CAATGCAAGACACCATCGTGG
Band lengths:
356-1518
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]