GgaEX6015397 @ galGal4
Exon Skipping
Gene
ENSGALG00000002546 | COL5A1
Description
collagen alpha-1(V) chain precursor [Source:RefSeq peptide;Acc:NP_990121]
Coordinates
chr17:7468273-7491836:+
Coord C1 exon
chr17:7468273-7468435
Coord A exon
chr17:7489682-7489813
Coord C2 exon
chr17:7491690-7491836
Length
132 bp
Sequences
Splice sites
3' ss Seq
TTTGGTTTCCCTCCCCGCAGGGC
3' ss Score
12.31
5' ss Seq
TACGTGAGT
5' ss Score
7.77
Exon sequences
Seq C1 exon
GTGGCACAGGGTCGCCATCAGCGTGCAGAAGAAAAACGTCACCTTGATTTTGGACTGTAAAAAGAAAATAACCAAGTTCTTGGATCGAAGCGACCATCCCATCATTGACGTCAATGGCATCATCGTATTTGGAACGAGGATATTAGATGAGGAAGTCTTCGAG
Seq A exon
GGCGACATCCAGCAGCTGCTGATCGTGGCCGACCCCCGGGCAGCCCACGACTACTGTGAGCACTACAGCCCCGACTGTGACACCGCCGTCCCCGATGCTCCACAATCCCAGGACCCCAACCAGGATGAATAC
Seq C2 exon
TATACCGATGGGGAGGGAGAAGGGGACACCTACTACTATGAGTACCCCTACTATGAAGACGTTGATGAAGCTGTAAAGCCTGAAGCACCCACCACCAAACCTGCACCCCCAGGAGTGGCTGCTGGAGAGAGACCCGAAACCAAGCAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSGALG00000002546-'3-3,'3-2,8-3=AN
Average complexity
A_S
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (Ref)
No structure available
Features
Disorder rate (Iupred):
C1=0.000 A=0.523 C2=0.837
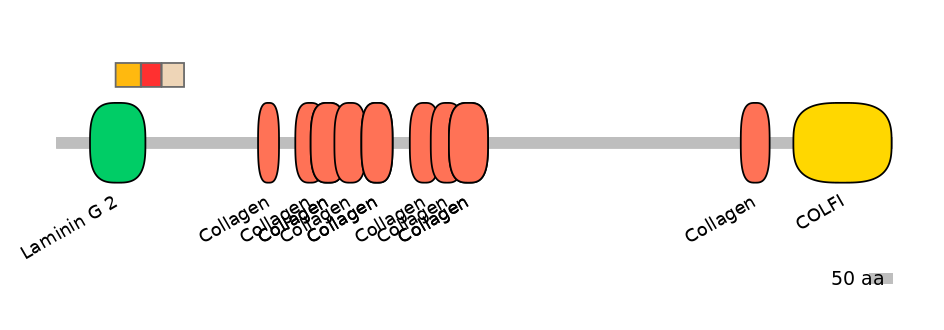
Domain overlap (PFAM):
C1:
PF0221019=Laminin_G_2=FE(45.0=100)
A:
PF0221019=Laminin_G_2=PD(7.5=20.5)
C2:
NO
Main Skipping Isoform:
NA
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Other assemblies
Conservation
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
GCCATCAGCGTGCAGAAGAAA
R:
GTGGGTGCTTCAGGCTTTACA
Band lengths:
242-374
Functional annotations
There are 1 annotated functions for this event
PMID: 12145749
[Not physiological]. Pro-alpha1(V) chains with abnormal N-propeptides were secreted and were incorporated into extracellular matrix, and the mutation resulted in dramatic alterations in collagen fibril structure. Note: also refers to a cryptic splice site.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]