HsaALTA0008403-2/2 @ hg38
Alternative 3'ss
Gene
ENSG00000197283 | SYNGAP1
Description
synaptic Ras GTPase activating protein 1 [Source:HGNC Symbol;Acc:HGNC:11497]
Coordinates
chr6:33446575-33447933:+
Coord C1 exon
chr6:33446575-33446786
Coord A exon
chr6:33447830-33447842
Coord C2 exon
chr6:33447843-33447933
Length
13 bp
Sequences
Splice sites
5' ss Seq
CAGGTGAGG
5' ss Score
10.07
3' ss Seq
CGCCACTAACCCCACTGAAGCCC
3' ss Score
-0.39
Exon sequences
Seq C1 exon
GTGAAGGAGTACGAGGAGGAGATTCACTCACTGAAAGAGCGGCTGCACATGTCCAACCGGAAGCTGGAAGAGTATGAGCGGAGGCTGCTGTCCCAGGAAGAACAAACCAGCAAAATCCTGATGCAGTATCAGGCCCGACTGGAGCAGAGTGAGAAGAGGCTAAGGCAGCAGCAGGCAGAGAAGGATTCCCAGATCAAGAGCATCATTGGCAG
Seq A exon
CCCGTCCCTTCAG
Seq C2 exon
GCTGATGCTGGTGGAGGAGGAGCTGCGCCGGGACCACCCCGCCATGGCTGAGCCGCTGCCAGAACCCAAGAAGAGGCTGCTCGACGCTCAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSG00000197283-59-58,59-57-2/2
Average complexity
Alt3
Mappability confidence:
NA
Protein Impact
ORF disruption when splice site is used (sequence inclusion)
No structure available
Features
Disorder rate (Iupred):
C1=0.744 A=NA C2=0.613
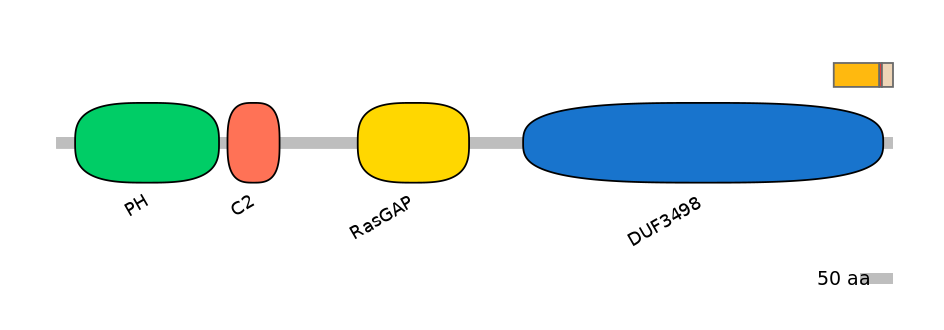
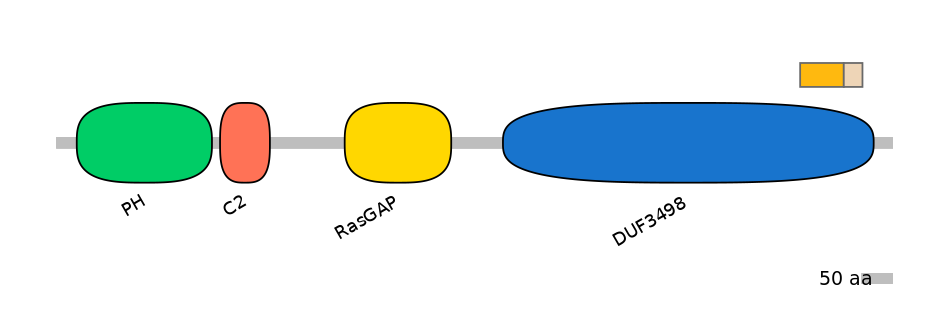
Domain overlap (PFAM):
C1:
PF120043=DUF3498=FE(12.1=100)
A:
NA
C2:
PF120043=DUF3498=FE(5.2=100)
Other Inclusion Isoforms:
NA
Associated events
Other assemblies
Conservation
Chicken
(galGal4)
No conservation detected
Chicken
(galGal3)
No conservation detected
Zebrafish
(danRer10)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
GGAGCAGAGTGAGAAGAGGCT
R:
CTCCTCCTCCACCAGCATCAG
Band lengths:
94-107
Functional annotations
There are 4 annotated functions for this event
PMID: 11278737
The beta isoform (13-nt frameshift from HsaALTA0008403), which lacks a C-terminal PSD-95-binding motif ((S/T)XV), was more restricted to the postsynaptic density fraction than the motif-containing alpha1 isoform. The beta isoform did not interact with PSD-95 but specifically interacted with a nonphosphorylated alpha subunit of Ca(2+)/calmodulin-dependent protein kinase II through its unique C-terminal tail.
PMID: 18824155
From the analysis using cultured neurons, unique expression of synGAP beta was found in a neuron with a sea urchin-like morphology, possibly a star pyramidal neuron, in which the synGAP beta expression was relatively high, in particular, at the distal part of its processes. SynGAP alpha1 was mostly or specifically localized to excitatory postsynaptic sites, whereas synGAP beta was present at both excitatory and inhibitory postsynaptic sites. Finally, there are more non-synaptic clusters in dendrites in the case of synGAP beta than synGAP alpha1.
PMID: 32068252
alpha1 isoforms were always found enriched in the postsynaptic density, alpha2 isoforms changed from a non-synaptic to a mostly postsynaptic density localization with age and beta isoforms were always found enriched in non-synaptic locations.
PMID: 32579114
SynGAP isoforms exhibit unique spatiotemporal expression patterns and play distinct roles in neuronal and synaptic development in mouse neurons. SynGAP-alpha1, which undergoes liquid-liquid phase separation with PSD-95, is highly enriched in synapses and is required for LTP. In contrast, SynGAP-beta (HsaALTA0008403, exon 18b), which does not bind PSD-95 PDZ domains, is less synaptically targeted and promotes dendritic arborization. A mutation in SynGAP-alpha1 that disrupts phase separation and synaptic targeting abolishes its ability to regulate plasticity and instead causes it to drive dendritic development like SynGAP-beta. Associated with APA usage.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- The Cancer Genome Atlas (TCGA)
- Autistic and control brains
- Pre-implantation embryo development