HsaEX0002148 @ hg38
Exon Skipping
Gene
ENSG00000072110 | ACTN1
Description
actinin alpha 1 [Source:HGNC Symbol;Acc:HGNC:163]
Coordinates
chr14:68877082-68880108:-
Coord C1 exon
chr14:68879962-68880108
Coord A exon
chr14:68878458-68878523
Coord C2 exon
chr14:68877082-68877240
Length
66 bp
Sequences
Splice sites
3' ss Seq
GGTACCACTGCCCCCCACAGAAG
3' ss Score
4.21
5' ss Seq
ATGGTAACG
5' ss Score
6.05
Exon sequences
Seq C1 exon
CACATCCGTGTGGGCTGGGAGCAGCTGCTCACCACCATCGCCAGGACCATCAATGAGGTAGAGAACCAGATCCTGACCCGGGATGCCAAGGGCATCAGCCAGGAGCAGATGAATGAGTTCCGGGCCTCCTTCAACCACTTTGACCGG
Seq A exon
AAGAAGACAGGCATGATGGACACGGATGATTTCCGCGCCTGCCTGATCTCCATGGGTTACAACATG
Seq C2 exon
GGAGAAGCAGAATTTGCCCGCATCATGAGCATTGTGGACCCCAACCGCCTGGGGGTAGTGACATTCCAGGCCTTCATTGACTTCATGTCCCGCGAGACAGCCGACACAGATACAGCAGACCAAGTCATGGCTTCCTTCAAGATCCTGGCTGGGGACAAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSG00000072110_MULTIEX2-2/2=C1-C2
Average complexity
ME(1-2[99=100])
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (No Ref)
No structure available
Features
Disorder rate (Iupred):
C1=0.081 A=0.023 C2=0.003
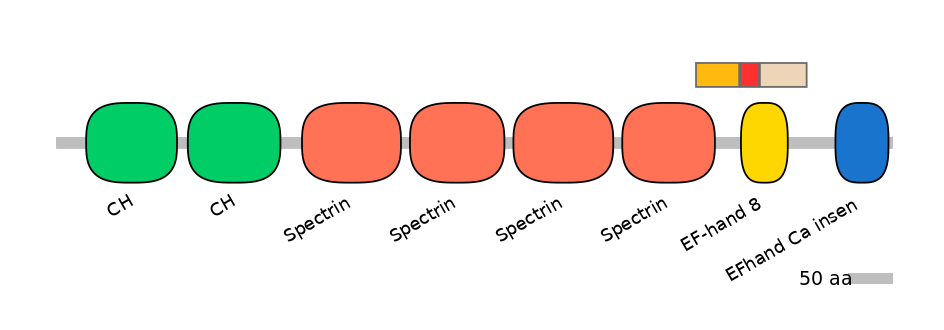
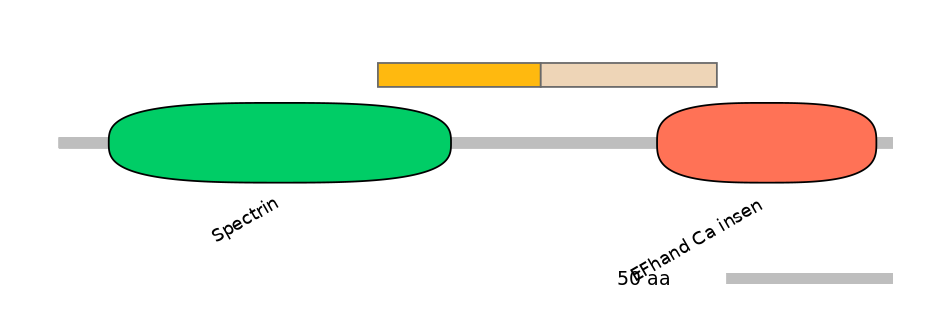
Domain overlap (PFAM):
C1:
PF0043516=Spectrin=PD(20.2=42.9),PF0003627=EF-hand_1=PU(34.5=20.4)
A:
PF138331=EF-hand_8=PU(37.7=90.9)
C2:
PF134991=EF-hand_7=PD(33.3=52.8),PF087265=EFhand_Ca_insen=PU(26.9=34.0)
Associated events
Other assemblies
Conservation
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
GGGCCTCCTTCAACCACTTTG
R:
GCCAGGATCTTGAAGGAAGCC
Band lengths:
175-241
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- The Cancer Genome Atlas (TCGA)
- Genotype-Tissue Expression Project (GTEx)
- Autistic and control brains
- Pre-implantation embryo development