HsaEX0061553 @ hg19
Exon Skipping
Gene
ENSG00000197694 | SPTAN1
Description
spectrin, alpha, non-erythrocytic 1 (alpha-fodrin) [Source:HGNC Symbol;Acc:11273]
Coordinates
chr9:131390204-131392654:+
Coord C1 exon
chr9:131390204-131390221
Coord A exon
chr9:131391404-131391466
Coord C2 exon
chr9:131392600-131392654
Length
63 bp
Sequences
Splice sites
3' ss Seq
GGTTTCTTTTCTTTGAATAGCAT
3' ss Score
6.37
5' ss Seq
AAGGTTTTT
5' ss Score
3.43
Exon sequences
Seq C1 exon
GACATACCTCCTCGATGG
Seq A exon
CATAGCATATCGTCGGGTCATTCGTGTCTATCAGTATGAAGTTGGGGATGATCTGTCTGGAAG
Seq C2 exon
GTCCTGTATGGTGGAAGAGTCGGGGACCCTCGAATCCCAGCTTGAAGCTACCAAA
VastDB Features
Vast-tools module Information
Secondary ID
ENSG00000197694_MULTIEX1-2/2=1-C2
Average complexity
C1
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (No Ref)
No structure available
Features
Disorder rate (Iupred):
C1=0.000 A=0.000 C2=0.561
Domain overlap (PFAM):
C1:
PF0043516=Spectrin=FE(20.7=100)
A:
PF0043516=Spectrin=PD(6.9=9.1),PF0043516=Spectrin=PU(4.8=18.2)
C2:
PF0043516=Spectrin=FE(17.0=100)
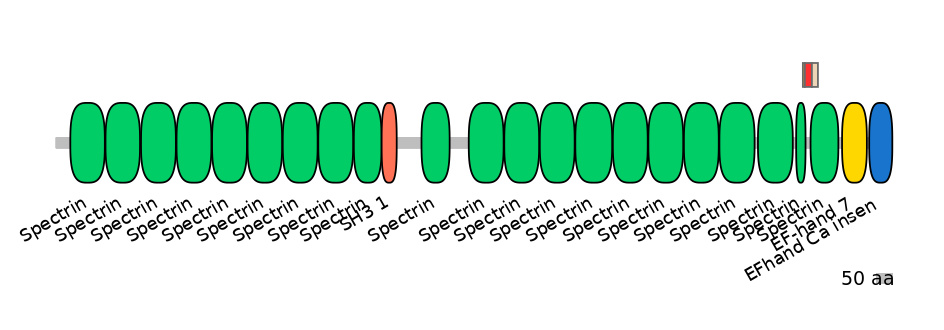
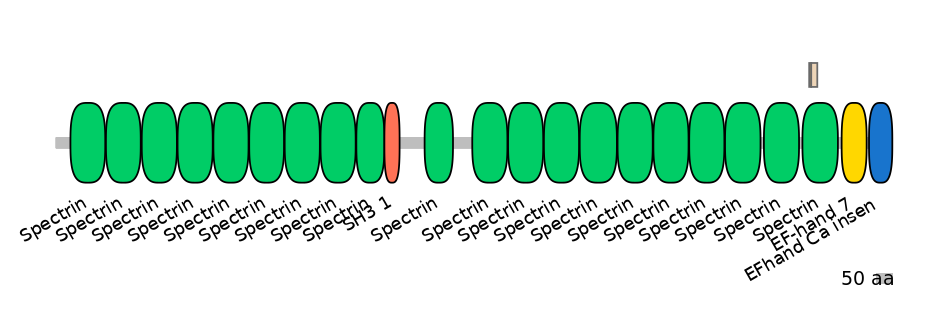
Other Inclusion Isoforms:
NA
Associated events
Other assemblies
Conservation
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
No suggested primer sequences
R:
No suggested primer sequences
Band lengths:
Functional annotations
There are 1 annotated functions for this event
PMID: 20114050
In vitro studies of recombinant alphaII-spectrin polypeptides representing the two splice variants of alphaII-spectrin, alphaII-cardi+ (with MmuEX0044726) and alphaII-cardi_ (without MmuEX0044726), revealed that the alphaII-cardi+ subunit has lower affinity for the complementary site in repeats 1-4 of betaII-spectrin, with a KD value of ~1 nM, as measured by surface plasmon resonance (SPR). In addition, the alphaII-cardi+ form showed 1.8-fold lower levels of binding to its site on betaII-spectrin than the alphaII-cardi_ form, both by SPR and blot overlay. This suggests that the 21-amino acid insert prevented some of the alphaII-cardi+ form from interacting with betaII-spectrin. Fusion proteins expressing the alphaII-cardi+ sequence within the two terminal spectrin repeats of alphaII-spectrin were insoluble in solution and aggregated in neonatal myocytes, consistent with the possibility that this insert removes a significant portion of the protein from the population that can bind beta subunits. Neonatal rat cardiomyocytes infected with adenovirus encoding GFP-fusion proteins of repeats 18-21 of alphaII-spectrin with the cardi+ insert formed many new processes. These processes were only rarely seen in myocytes expressing the fusion protein lacking the insert or in controls expressing only GFP. These results suggest that the embryonic mammalian heart expresses a significant amount of alphaII-spectrin with a reduced avidity for beta-spectrin and the ability to promote myocyte growth.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- The Cancer Genome Atlas (TCGA)
- Genotype-Tissue Expression Project (GTEx)
- Autistic and control brains
- Pre-implantation embryo development
Other AS DBs:
FasterDB (Includes CLIP-seq data)
AS-ALPS (AS-induced ALteration of Protein Structure, links to PINs)
APPRIS (Selection of principal isoform)
DEU primates (Only for human)