MmuEX6045151 @ mm9
Exon Skipping
Gene
ENSMUSG00000035232 | Pdk3
Description
pyruvate dehydrogenase kinase, isoenzyme 3 [Source:MGI Symbol;Acc:MGI:2384308]
Coordinates
chrX:91045453-91054281:-
Coord C1 exon
chrX:91054210-91054281
Coord A exon
chrX:91047548-91047732
Coord C2 exon
chrX:91045453-91045542
Length
185 bp
Sequences
Splice sites
3' ss Seq
TGTATTTCCTTCCCACACAGCTT
3' ss Score
8.03
5' ss Seq
ACAGTAAGT
5' ss Score
9.49
Exon sequences
Seq C1 exon
GTACATGCAGAGCTTCCTTGAACTTTTAGAATATGAAAACAAGAGCCCAGAAGACCCACGAGTTTTGGATAA
Seq A exon
CTTTCTCAACGTTCTGATAAATATCAGAAACAGACACAATGATGTTGTTCCTACAATGGCCCAAGGCGTGATTGAGTACAAGGAAAAGTTCGGGTTTGATCCGTTCATTAGCAGTAACATTCAATATTTCCTGGATCGGTTTTATACCAACCGCATTTCTTTCCGCATGCTTATTAACCAGCACA
Seq C2 exon
CACTTCTGTTTGGTGGTGACACTAACCCTGCACATCCGAAACATATAGGGAGTATCGACCCCACCTGTAATGTAGCTGATGTGGTTAAAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSMUSG00000035232-'4-3,'4-2,5-3=AN
Average complexity
A_S
Mappability confidence:
100%=100=100%
Protein Impact
ORF disruption upon sequence exclusion
No structure available
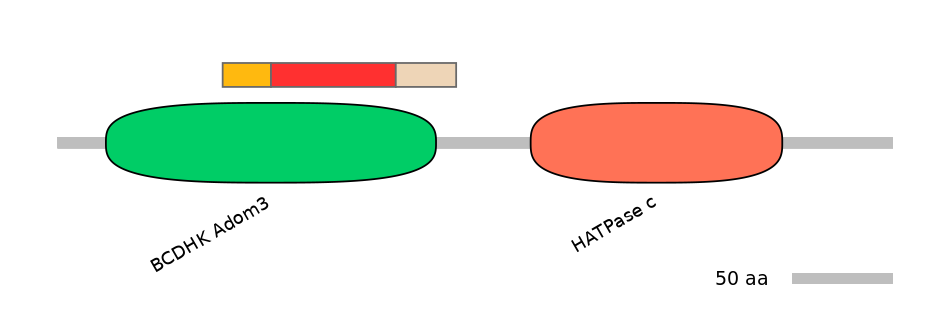
Features
Disorder rate (Iupred):
C1=0.000 A=0.000 C2=0.065
Domain overlap (PFAM):
C1:
PF104364=BCDHK_Adom3=FE(14.5=100)
A:
PF104364=BCDHK_Adom3=FE(37.6=100)
C2:
PF104364=BCDHK_Adom3=PD(12.1=64.5)
Main Skipping Isoform:
NA
Other Skipping Isoforms:
NA
Associated events
Other assemblies
Conservation
Primers PCR
Suggestions for RT-PCR validation
F:
GTACATGCAGAGCTTCCTTGA
R:
CTTTAACCACATCAGCTACATTACA
Band lengths:
162-347
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
Other AS DBs: