MmuEX6100701 @ mm9
Exon Skipping
Gene
ENSMUSG00000038740 | Fam125b
Description
family with sequence similarity 125, member B [Source:MGI Symbol;Acc:MGI:1919793]
Coordinates
chr2:33680568-33690153:-
Coord C1 exon
chr2:33690057-33690153
Coord A exon
chr2:33683202-33683331
Coord C2 exon
chr2:33680568-33680690
Length
130 bp
Sequences
Splice sites
3' ss Seq
TTCTATCCTCCTTTCCTTAGAGG
3' ss Score
9.2
5' ss Seq
CGGGTAAGT
5' ss Score
10.75
Exon sequences
Seq C1 exon
AGTCATTTGGGGAATGTGCTGGTGGACATGAAGCTCATCGATGTCAAGGACACATTGCCTGTGGGCTTCATCCCCATCCAAGAGACCGTGGATACAC
Seq A exon
AGGAGGTGGTCTTTAGGAAGAAGCGGCTCTGCATTAAGTTTATCCCCCGGGATTCAACTGAAGCTGCCATCTGTGACATTCGCATCATGGGCCGGACCAAGCAGGCACCGCCCCAGTACACGTTTATCGG
Seq C2 exon
GGAACTGAACAGCATGGGCATCTGGTACCGAATGGGCAGAGTGCCAAGAAACCATGACTCGTCTCAGCCCACCACGCCCTCCCAGTCATCAGCCTCTTCTACCCCAGCCCCAAACCTTCCCAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSMUSG00000038740-'11-12,'11-10,12-12=AN
Average complexity
A_S
Mappability confidence:
100%=100=100%
Protein Impact
ORF disruption upon sequence exclusion
No structure available
Features
Disorder rate (Iupred):
C1=0.000 A=0.000 C2=0.600
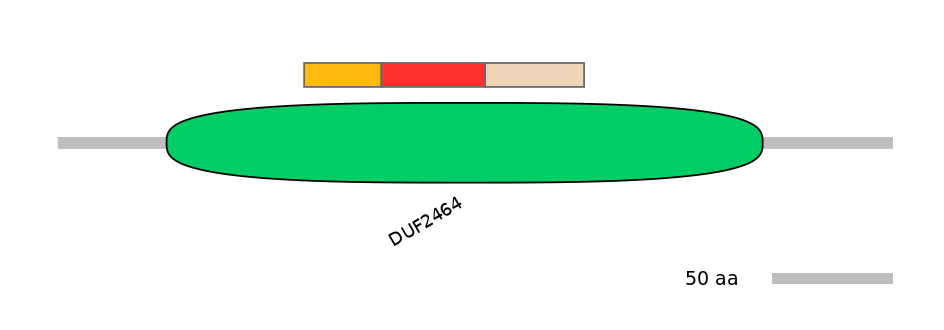
Domain overlap (PFAM):
C1:
PF102404=DUF2464=FE(12.7=100)
A:
PF102404=DUF2464=FE(17.1=100)
C2:
PF102404=DUF2464=PD(23.1=95.2)
Main Skipping Isoform:
NA
Other Skipping Isoforms:
NA
Associated events
Other assemblies
Conservation
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
TCATTTGGGGAATGTGCTGGT
R:
CTGGGAAGGTTTGGGGCTG
Band lengths:
218-348
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
Other AS DBs: