RnoEX0036156 @ rn6
Exon Skipping
Gene
ENSRNOG00000058359 | Fkbp8
Description
FK506 binding protein 8 [Source:RGD Symbol;Acc:1308670]
Coordinates
chr16:20649605-20652889:-
Coord C1 exon
chr16:20652852-20652889
Coord A exon
chr16:20651060-20651346
Coord C2 exon
chr16:20649605-20649774
Length
287 bp
Sequences
Splice sites
3' ss Seq
ATGCTTAACTGTTCCCCTAGGGC
3' ss Score
8.59
5' ss Seq
TGGGTAAGT
5' ss Score
10.24
Exon sequences
Seq C1 exon
TGGGGGTGGGGAGGGGGGCACAGGACCCCAAGCCGCAG
Seq A exon
GGCCAGTTCCTGATCCCCCAGCAGCATGGCGTCTTGGGCCGAGCCCTCTGAGCCTGCTGCCCAGCTACTTTGTGGGGCCCCACTCCTGGAGGGCTTTGAGGTGCTTGATGGGGTTGATGATGCAGAGGAGGAAGATGACCTAAGTGGGTTGCCACCACTAGAGGATATGGGACAGCCTACGGTGGAGGAGGCTGAGCAGCCTGGAGCCCTAGCTCGGGAGTTCCTGGCAGCCACAGAGCCTGAGCCAGCCCCAGCCCCCGCCCCGGAAGAGTGGCTGGACATTCTGG
Seq C2 exon
GGAACGGATTGCTGCGTAAGAAGACACTGGTCCCAGGCCCAACAGGCTCTAGCCGCCCACTCAAGGGCCAGGTGGTGACCGTACACCTGCAGATGTCCCTGGAGAATGGCACCCGTGTGCAAGAGGAGCCCGAACTGGCTTTCACGCTTGGAGACTGCGATGTTATCCAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSRNOG00000058359-'3-2,'3-0,13-2
Average complexity
C1
Mappability confidence:
94%=100=80%
Protein Impact
ORF disruption upon sequence exclusion (Ref, Alt. ATG (<=10 exons))
No structure available
Features
Disorder rate (Iupred):
C1=NA A=0.830 C2=0.474
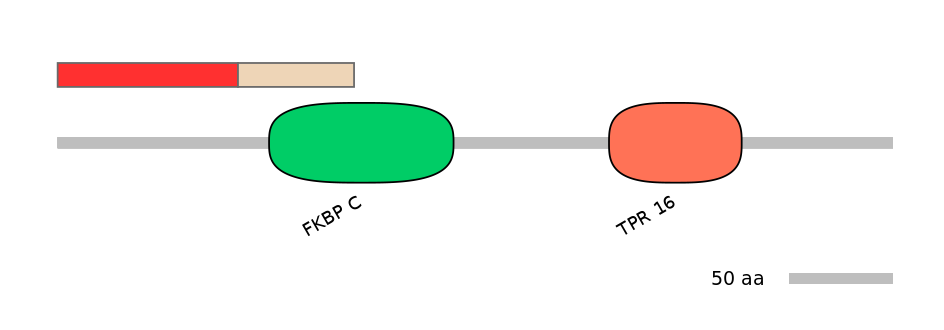
Domain overlap (PFAM):
C1:
NA
A:
NO
C2:
PF0025423=FKBP_C=PU(45.6=71.9)
Main Skipping Isoform:
NA
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Conservation
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
No suggested primer sequences
R:
No suggested primer sequences
Band lengths:
Functional annotations
There are 2 annotated functions for this event
PMID: 19546213
Encodes an experimentally validated Eukaryotic Linear Motif (ELM). Method: affinity chromatography technology, anti tag coimmunoprecipitation, colocalization, competition binding, detection by mass spectrometry, fluorescence polarization spectroscopy, fluorescent resonance energy transfer, glutathione s tranferase tag, isothermal titration calorimetry, mutation analysis, protein array, pull down, subcellular preparation. ELM ID: ELMI002479; ELM sequence: LPPLE; Overlap: FULL
PMID: 23413029
Encodes an experimentally validated Eukaryotic Linear Motif (ELM). Method: affinity chromatography technology, anti tag coimmunoprecipitation, colocalization, competition binding, detection by mass spectrometry, fluorescence polarization spectroscopy, fluorescent resonance energy transfer, glutathione s tranferase tag, isothermal titration calorimetry, mutation analysis, protein array, pull down, subcellular preparation. ELM ID: ELMI002479; ELM sequence: LPPLE; Overlap: FULL
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]