RnoINT0132328 @ rn6
Intron Retention
Gene
ENSRNOG00000005229 | Sec16b
Description
SEC16 homolog B, endoplasmic reticulum export factor [Source:RGD Symbol;Acc:621658]
Coordinates
chr13:75184636-75185203:+
Coord C1 exon
chr13:75184636-75184787
Coord A exon
chr13:75184788-75185141
Coord C2 exon
chr13:75185142-75185203
Length
354 bp
Sequences
Splice sites
5' ss Seq
GAGGTAATC
5' ss Score
7.05
3' ss Seq
AATTTTATTTTCTACTCCAGGTT
3' ss Score
9.23
Exon sequences
Seq C1 exon
ATGACACCTCAGCGACCGTTCCCAAGGCACCCATGAGGTTCTACGTTCCCCACGTGTCTGTGAGCTTTGGGCCAGGAGGCCAGCTGATATGTGTGTCCCCCAACTCTCCTGCTGATGGACAAACGGCCCTTGTTGAAGTGCACAGCATGGAG
Seq A exon
GTAATCAATATGGTGGGCCTCGGAGATGCAAGTCCTCTGCTCAATTCAACCGCTCAGCTCACTTGCACAGGCTTTCACTTACTTTGCTTGAGGCTGGGAGTCTCCAGGAGGGGGAATTGAATTACGTGTGCCCTAGATTGTGGGCAAGAATGGGTGGGCTTTTCTCTGAAGGAAGTGATGATGCTAGATTTACATTTTCCCTGAGATGCTAGTGACTCATCTTTCAACAGGATAACTCTCCCCACAAGGCACTGAGCCACACCATAAAACCAAAGCCCTGGATGTGAGACTCCATTCTACTGGTGGGTACTGGCTGGAAGAATCTTGAGGACCTAATTTTATTTTCTACTCCAG
Seq C2 exon
GTTATACTTAATGATTTTGAGGATCAAGAGGAGATGAGAGCTTTCCCAGGGCCCCTCATCAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSRNOG00000005229:ENSRNOT00000007321:6
Average complexity
IR
Mappability confidence:
NA
Protein Impact
ORF disruption upon sequence inclusion
No structure available
Features
Disorder rate (Iupred):
C1=0.157 A=NA C2=0.000
Domain overlap (PFAM):
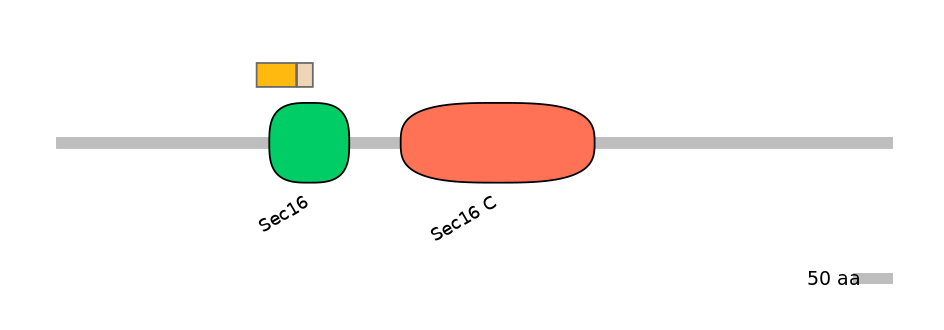
C1:
PF129322=Sec16=PU(33.3=66.7)
A:
NA
C2:
PF129322=Sec16=FE(19.6=100)
Main Inclusion Isoform:
NA
Other Inclusion Isoforms:
NA
Associated events
Conservation
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
ATGACACCTCAGCGACCGTTC
R:
CTGATGAGGGGCCCTGGG
Band lengths:
214-568
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]