DmeEX0005781 @ dm6
Exon Skipping
Gene
FBgn0020621 | Pkn
Description
The gene Protein kinase N is referred to in FlyBase by the symbol DmelPkn (CG2049, FBgn0020621). It is a protein_coding_gene from Dmel. It has 17 annotated transcripts and 17 polypeptides (14 unique). Gene sequence location is 2R:9252310..9285097. Its molecular function is described by: protein serine/threonine kinase activity; protein kinase activity; ATP binding; protein binding; GTP-Rho binding. It is involved in the biological process described with 9 unique terms, many of which group under: cellular component organization; regulation of wound healing; localization; cell division; post-embryonic animal organ development. 65 alleles are reported. The phenotypes of these alleles manifest in: cellular anatomical entity; array of sensillum rows; non-connected developing system; adult head; intracellular non-membrane-bounded organelle. The phenotypic classes of alleles include: female semi-sterile; some die during larval stage; increased mortality; phenotype. Summary of modENCODE Temporal Expression Profile: Temporal profile ranges from a peak of moderately high expression to a trough of moderate expression. Peak expression observed at stages throughout embryogenesis, during late larval stages, at stages throughout the pupal period, in adult female stages.
Coordinates
chr2R:9267655-9284129:-
Coord C1 exon
chr2R:9283828-9284129
Coord A exon
chr2R:9271985-9272174
Coord C2 exon
chr2R:9267655-9267976
Length
190 bp
Sequences
Splice sites
3' ss Seq
ATTTTCGCATATTTCTACAGTGA
3' ss Score
6.59
5' ss Seq
CACGTATGT
5' ss Score
7.26
Exon sequences
Seq C1 exon
GAAATCAAATGAGAGCGCATTCGATAAAAAGAGATAAAGAAAATACTGTTCGGCATCAGGTGTTAAAGTGCTAAACAATTCCGAAAGCCAAAGGAAAGTAAACAAAAACGCGACGCGTGTGCTTGTGCTTGTGTGCGGCGCCAATAAAAGCCAAAAGCAAAATACTGACAACCAAAAAGATACACGCAACAACAAGAACCATATTCATAATAACCAACAACTAAATACTACAACAAGAAGCTACGGCATCGAGTGATTCAACGCGGCGCACCAAATAAACCATGTCGGATTCGTATTATCAG
Seq A exon
TGAGCTAACGAAAAGAAAGAAATTAACTTTAAATATACTGCCAATGTGTTACTATCGGTGTACGAATATCACATTCAATGCCGTTTGTGTGAAATAAACTAAGCAAATAGAAGAAACCCGTACTATGGCATTGACCATGAACATGGTTTTCCTCAAGGACCTACGATCGCGCCTCAAGGGATATCTGCAC
Seq C2 exon
GGCGAATACATCAAGCATCCCGTTCTGTACGAGCTCAGTCACAAATATGGTTTCACAGAGAATCTGCCGGAGAGCTGTATGTCCATACGGCTGGAGGAGATCAAGGAGGCCATTCGGCGAGAGATCCGCAAGGAGCTGAAGATCAAAGAGGGCGCCGAGAAGCTCCGCGAGGTGGCTAAAGATCGACGATCCCTCAGCGATGTGGCCGTTCTTGTCAAGAAGAGCAAAAGTAAACTTGCCGAGCTGAAGTCCGAGTTGCAGGAGCTCGAGAGTCAAATCCTCCTGACATCGGCCAACACCGCCGTCAATAGTAATGGACAAG
VastDB Features
Vast-tools module Information
Secondary ID
FBgn0020621_MULTIEX2-2/3=C1-C2
Average complexity
S
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (No Ref, Alt. ATG)
No structure available
Features
Disorder rate (Iupred):
C1=0.000 A=0.000 C2=0.000
Domain overlap (PFAM):
C1:
NO
A:
NO
C2:
PF0218511=HR1=WD(100=62.0)
Main Inclusion Isoform:
FBpp0087685
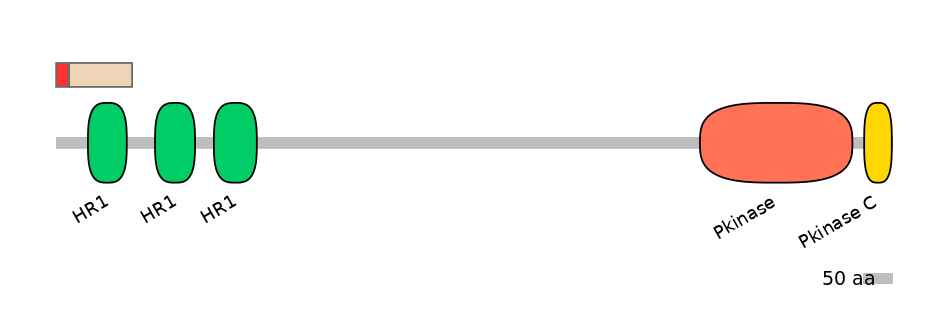
Main Skipping Isoform:
FBpp0308399
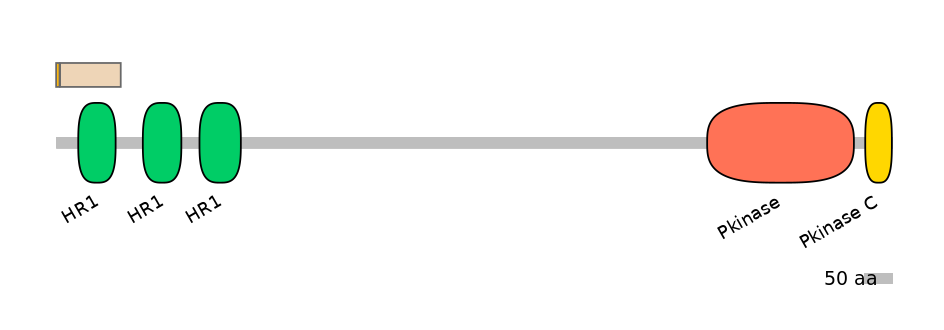
Other Inclusion Isoforms:
FBpp0087684, FBpp0291759, FBpp0297540
Other Skipping Isoforms:
FBpp0087682, FBpp0087683, FBpp0300526, FBpp0308400, FBpp0308401, FBpp0308402
Associated events
Conservation
Chicken
(galGal4)
No conservation detected
Chicken
(galGal3)
No conservation detected
Zebrafish
(danRer10)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
AGAAGCTACGGCATCGAGTGA
R:
GTCGATCTTTAGCCACCTCGC
Band lengths:
254-444
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Neural diversity
- Neurogenesis
- Neuronal activity
- Splicing factor regulation (brain)
- Splicing factor regulation (SL2)