DmeEX6002213 @ dm6
Exon Skipping
Gene
FBgn0030503 | Tango2
Description
The gene Transport and Golgi organization 2 is referred to in FlyBase by the symbol DmelTango2 (CG11176, FBgn0030503). It is a protein_coding_gene from Dmel. It has 5 annotated transcripts and 5 polypeptides (2 unique). Gene sequence location is X:13616989..13619207. Its molecular function is described by: . It is involved in the biological process described with: Golgi organization; protein secretion. 5 alleles are reported. No phenotypic data is available. The phenotypic class of alleles includes: viable. Summary of modENCODE Temporal Expression Profile: Temporal profile ranges from a peak of high expression to a trough of moderate expression. Peak expression observed during early pupal stages.
Coordinates
chrX:13617814-13619203:+
Coord C1 exon
chrX:13617814-13617936
Coord A exon
chrX:13618273-13618402
Coord C2 exon
chrX:13618597-13619203
Length
130 bp
Sequences
Splice sites
3' ss Seq
TAACGAACATTTCTTACTAGGGC
3' ss Score
5.44
5' ss Seq
TGGGTGAGT
5' ss Score
8.73
Exon sequences
Seq C1 exon
GGATCGATCTGGAGCCGGGACGCGAGGGCGGCACCTGGCTGGCGATCGGCCACTCCGCTGGCTTCTTTAAGGTGGGCGCCCTGCTCAATCTGACCGGCGAGCCCAAGCCACGCGATGCAGTTG
Seq A exon
GGCGCGGCATGATTGTGGCGGACTATGTTACCCGGGCGGACGAGGAGCACAGCATCCTGAACTACAACGAAAGGCTGCTCAAAGATTGCACCAAGTATTCTGCCTTCAACTTTGTGTCCATCGAAATTGG
Seq C2 exon
ATCTGCATCCCAGCCCGCTCGCGTGAAGCTGCTGAGCAATGTGCCGCCCACGCTGGAGGATTTCCAGAACGGCGAGTGCTACGGATTTGGCAACAGTCTGCCGCACTCTCCGTTCGAGAAGGTGCGTCATGGCAAACAGGAGTTCGAGGCGATCGTTAAGGCTCATGGGGAAGCCAGCGTGGAGACTCTGTCCGCCCAGCTGATGCAGTTGCTCCGCAACAAGCACAAATTCTGGCCGGACGACGAGCTAAAGACACGTGCGCCCAACTGGGGCGAGGGATTGAGTTCCCTAAACGTCCATATCGAAGAGCATGCCTATGGCAGCCGTACACACTCCGTTGTCCTGGTGGACAGCGAGAATAAGATGCACTTCATTGAGGAGACAATGACTGGATTGGATCCGCACGGCGAATGGAACAAGACCCACATTGAAAAGGATTTTCAAAACGGCGTTTGACCACGGACAGGATGACGCAACTTCTTGACGCAACTATCTCGTT
VastDB Features
Vast-tools module Information
Secondary ID
FBgn0030503-'8-8,'8-6,9-8=AN
Average complexity
A_S
Mappability confidence:
100%=100=100%
Protein Impact
ORF disruption upon sequence exclusion
No structure available
Features
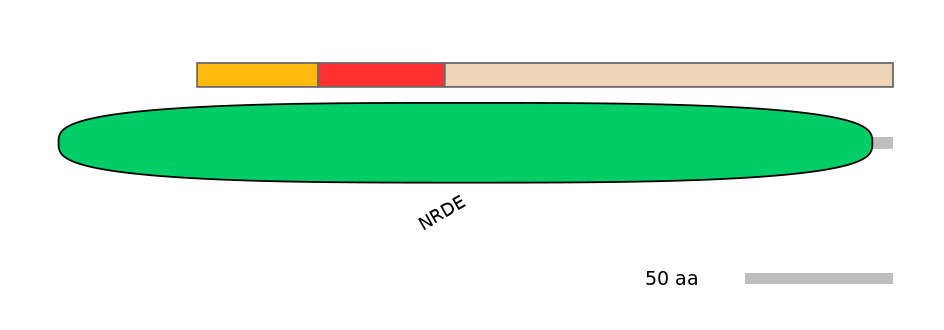
Disorder rate (Iupred):
C1=0.048 A=0.223 C2=0.197
Domain overlap (PFAM):
C1:
PF057427=NRDE=FE(14.8=100)
A:
PF057427=NRDE=FE(15.5=100)
C2:
PF057427=NRDE=PD(52.3=94.8)
Main Inclusion Isoform:
FBpp0073638
Main Skipping Isoform:
NA
Other Inclusion Isoforms:
FBpp0300867, FBpp0304111, FBpp0309341
Other Skipping Isoforms:
NA
Associated events
Conservation
Primers PCR
Suggestions for RT-PCR validation
F:
TCCGCTGGCTTCTTTAAGGTG
R:
GGCTTCCCCATGAGCCTTAAC
Band lengths:
245-375
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Neural diversity
- Neurogenesis
- Neuronal activity
- Splicing factor regulation (brain)
- Splicing factor regulation (SL2)