DmeEX6014489 @ dm6
Exon Skipping
Gene
FBgn0035449 | CG14971
Description
This gene is referred to in FlyBase by the symbol DmelCG14971 (FBgn0035449). It is a protein_coding_gene from Dmel. It has 2 annotated transcripts and 2 polypeptides (1 unique). Gene sequence location is 3L:3468023..3471309. Its molecular function is described by: transmembrane transporter activity; antiporter activity. It is involved in the biological process described with: UDP-glucose transmembrane transport. 7 alleles are reported. No phenotypic data is available. The phenotypic classes of alleles include: fertile; viable. Summary of modENCODE Temporal Expression Profile: Temporal profile ranges from a peak of moderately high expression to a trough of low expression. Peak expression observed within 00-06 hour embryonic stages, during late larval stages, during early pupal stages.
Coordinates
chr3L:3468023-3471028:-
Coord C1 exon
chr3L:3470388-3471028
Coord A exon
chr3L:3469383-3469635
Coord C2 exon
chr3L:3468023-3469322
Length
253 bp
Sequences
Splice sites
3' ss Seq
ATGGCATTATTTACTTTCAGAGC
3' ss Score
6.54
5' ss Seq
AAGGTTAGT
5' ss Score
8.54
Exon sequences
Seq C1 exon
CGCAGCTGGGTAATGGCCATCGGTTGCGCCAGAGCAACTTCAAGTACGCCAGCGGAAAGTGCGGATCCGGAGAGGCGCCGTGCACCAACGAAACGGCCACGTTGGCCCAGGAGAACATGATGATGCAAATGGCCGTGGGTACGCTGGCCATTATCTTCCTGTATCTGGCATTATCCATATCGCTGACATTCTACCAAACGGACATCAACCGGCAGATGCCCTTTCCCCTGGCCATCGTAACCTATCATTTGGTTGTGAAGTTCCTTCTGGCCGCCGCGGCGCGCAGAATCTACAGGATGCGAGTGGGCCGATCGCGGGTTCAGTTGGATTGGAGGCTCGCCCTGCGGAAAATGGCACCCACGGGAGTTGCCAGTGCCATCGACATTGGATTCTCCAACTGGGGCCTGGCCCTGGTGCCCATTTCCCTCTACACCATGACCAAGTCCTCGACCATTGTTTTTATCCTGCTGTTCGCCATCGCTTTTGGACTGGAGAAGAAG
Seq A exon
AGCTGGTATCTGGTCTCGATTGTGGGCCTGATAGGCACTGGCCTTCTGATGTTCACCTACAAGTCAACGGACTTCAATGCCCTTGGTTTCTTCTTCATCCTTTTCGCCTCGCTGAGCAGCGGACTGCGATGGAGTTTCGCGCAGTTCATAATGCAGAAATCGAAGCTAGGCTTGCACAATCCCATCGATATGATATATTACATGCAGCCGTGGATGATTGCCTCGCTGGTGCCCCTCGTAATTGGAATCGAAG
Seq C2 exon
GTGCCGGGCTCATCGCGGTAATAGAAGATCTGCACAACCACACTTCGAATGAGATAACTTGGGCCATTGCGAGGATATCAGCGGGCGCTTTGCTGGCTTTTCTCATGGAGTTCAGCGAATTCTTGGTGCTCTGCAAGACCTCCAGTCTGACGCTCTCGATAGCCGGCATTTTCAAGGATATCTGCCAGTTGGCACTAGCCGTGACTATCAGAAAAGACCACCTGAGTGTCATCAACTACATTGGCTTGATCATTTGCCTGGCTGGTATCGTTTGCCACCTACTGCACAAGTACTCCAATATGAAGGAGATGCAGCGGCAGCAGGAGTTGCAGCTCGATAACGACCAGGAGGAGTCGTCGCCAGGGGAGTACAAATTCAACGATGGCAGTGCCATTGTGGGTGTCCATGTGAAGTCGCACTCCACGCTGACGGTGCCATTGCTGGAGCAAACCGACTCGGAGGACGAGTCTGGTGCAAGCAACAAACAGAACGCCTCCGAT
VastDB Features
Vast-tools module Information
Secondary ID
FBgn0035449-'1-4,'1-3,6-4=AN
Average complexity
A_S
Mappability confidence:
100%=100=100%
Protein Impact
ORF disruption upon sequence exclusion
No structure available
Features
Disorder rate (Iupred):
C1=0.112 A=0.000 C2=0.167
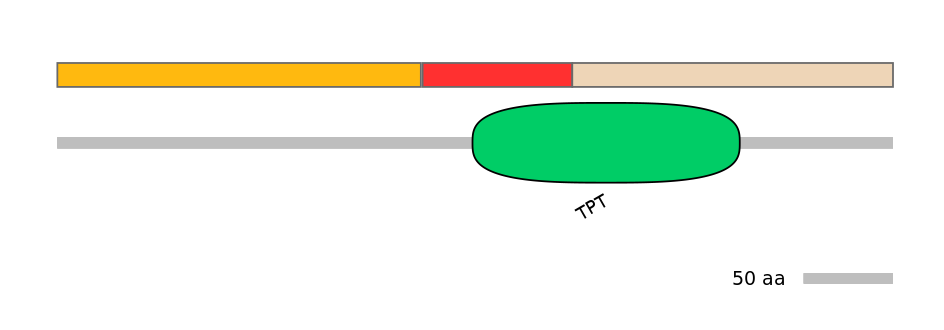
Domain overlap (PFAM):
C1:
NO
A:
PF0315111=TPT=PU(37.1=65.9)
C2:
PF0315111=TPT=PD(62.3=51.9)
Main Inclusion Isoform:
FBpp0073002
Main Skipping Isoform:
NA
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Conservation
Primers PCR
Suggestions for RT-PCR validation
F:
CCAGTGCCATCGACATTGGAT
R:
AAAATGCCGGCTATCGAGAGC
Band lengths:
303-556
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Neural diversity
- Neurogenesis
- Neuronal activity
- Splicing factor regulation (brain)
- Splicing factor regulation (SL2)