DreEX0078492 @ danRer10
Exon Skipping
Gene
ENSDARG00000017494 | tgfbr1a
Description
transforming growth factor, beta receptor 1 a [Source:ZFIN;Acc:ZDB-GENE-051120-75]
Coordinates
chr2:24221134-24234624:-
Coord C1 exon
chr2:24234391-24234624
Coord A exon
chr2:24224657-24224896
Coord C2 exon
chr2:24221134-24221364
Length
240 bp
Sequences
Splice sites
3' ss Seq
TTTGATTCTACCTGTTTCAGAGT
3' ss Score
7.53
5' ss Seq
CAGGTACTG
5' ss Score
9.04
Exon sequences
Seq C1 exon
GACTGCTGTGTTACTGTGAACGATGTGTCAATCGGTCGTGCAACACAACTGGACTGTGTTTTGCCGTCATCGCTAAGTCTTCTGGACAGACCGTGACACAGGAGCATCAGTGTATTCAGCAGGATGATCTTTACCCCCGGGACAGGCCTTTCCAGTGCGCTCCTTCCAACAACGGGAAGCATCCGTACTGCTGCAACACTCACATGTGCAACAAGAACCCAAAAGTTATCCCAG
Seq A exon
AGTTGCCTGAAGCTGAACCCCAGTCTATGAGTCCCATAGCCTTGGCCGCAATGATTGCTGTCCCCATCTGTGTGCTGAGCTTTGTGCTGGTGCTGCTTTTCTACATGTGCCATAATCGCTCAATCATCCACCATCGTGTGCCAAGTGAAGAAGACCCTACTATGGACCACCCATTCCTTGCTGATGGCACCACACTTAAGGACCTCATCTATGACATGACCACCTCTGGATCAGGCTCAG
Seq C2 exon
GTTTGCCTCTGCTAGTGCAACGGACCATCGCCAGGACCATCATCCTCCAAGAGAGCATTGGGAAGGGCCGTTTTGGAGAGGTGTGGAGAGGCCGCTGGCGTGGAGAGGAGGTGGCGGTGAAGATATTCTCCTCCAGAGAGGAGCGCTCTTGGTTCCGCGAGGCAGAAATCTACCAGACTGTCATGCTCCGTCATGAGAACATCCTGGGCTTTATTGCTGCAGACAACAAAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSDARG00000017494_CASSETTE1
Average complexity
S
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (Ref)
No structure available
Features
Disorder rate (Iupred):
C1=0.000 A=0.148 C2=0.000
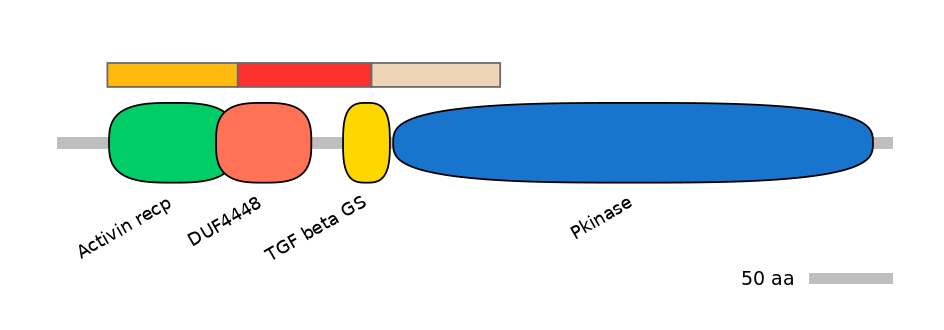
Domain overlap (PFAM):
C1:
PF0106418=Activin_recp=WD(100=97.5),PF146101=DUF4448=PU(22.4=16.5)
A:
PF146101=DUF4448=PD(75.9=54.3),PF085157=TGF_beta_GS=PU(58.6=21.0)
C2:
PF085157=TGF_beta_GS=PD(37.9=14.1),PF0006920=Pkinase=PU(22.2=82.1)
Main Skipping Isoform:
NA
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Conservation
Primers PCR
Suggestions for RT-PCR validation
F:
ACGATGTGTCAATCGGTCGTG
R:
CTCCACACCTCTCCAAAACGG
Band lengths:
303-543
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]